MOJ
eISSN: 2374-6920


Research Article Volume 2 Issue 1
Department of Biological Sciences, Charles E. Schmidt College of Science, Florida Atlantic University, USA
Correspondence: Ramaswamy Narayanan, Department of Biological Sciences, Charles E. Schmidt College of Science, Florida Atlantic University, FL 33431, USA, Tel 15612972247, Fax +15612973859,
Received: January 08, 2015 | Published: January 30, 2015
Citation: Delgado AP, Chapado MJ, Brandao P, et al. Atlas of the open reading frames in human diseases: dark matter of the human genome. MOJ Proteomics Bioinform. 2015;2(1):16-25. DOI: 10.15406/mojpb.2015.02.00036
Background: The human genome encodes RNA and protein coding sequences, the non-coding RNAs, pseudogenes and uncharacterized Open Reading Frames (ORFs). The dark matter of the human genome encompassing the uncharacterized proteins and the non-coding RNAs are least well studied. However, they offer novel druggable targets and biomarkers discovery for diverse diseases.
Methods: In this study, we have systematically dissected the dark matter of the human genome. Using diverse bioinformatics tools, an atlas of the dark matter of the genome was created. The ORFs were characterized for gene ontology, mRNA and protein expression, protein motif and domains and genome-wide association.
Results: An atlas of disease-related ORFs (n=800) was generated. A complex landscape of involvement in multiple diseases was seen for these ORFs. Motif and domains analysis identified druggable targets and putative secreted biomarkers including enzymes, receptors and transporters as well as proteins with signal peptide sequence. About ten percent of the ORFs showed a correlation of gene expression at the mRNA and protein levels. Genome-Phenome association tools identified ORF’s association with autoimmune, cardiovascular, cancer, diabetes, infection, metabolic, neurodegenerative and psychiatric disorders. Tumors encompassed largely heterozygous mutations.
Conclusions: The dark matter human proteome atlas reported in this study should aid in the discovery of novel therapeutic and diagnostic targets for multiple diseases.
Keywords: bioinformatics, biomarkers, body fluids, breast cancer, cancer proteome, dark matter, druggable targets, human genome, open reading frames, pancreatic cancer, prostate cancer, uncharacterized proteins
ClinVar, clinical variations; COSMIC, catalogue of somatic mutations in cancer; eQTL, expression quantitative trait loci; EBI-EMBL, european bioinformatics institute- european molecular biology laboratory; GAD, genetic association database; GtEx, genotype-tissue nomenclature committee; HapMap, haploid map; HGNC, human genome nomenclature committee; HPRD, human protein reference database; HPA, human protein atlas; MOPED, model organism protein expression database; PheGenI, phenotype-genotype integrator; ORF, open reading frame; RefSeq, reference sequence; SNP, single nucleotide polymorphism; TCGA, cancer gene atlas; RP, retinitis pigmentosa; CRD, cone-rod dystrophy; AI, amelogenesis imperfecta; JBTS, joubert syndrome; FTD, frontotemporal dementia; ALS, amyotrophic lateral sclerosis; ORFs, open reading frames
A vast amount of the human genome still remains uncharacterized.1–4 The uncharacterized part of the human genome offers an attractive potential for novel target discovery for diverse diseases.5 While known genes are easier to follow in the laboratory, a gap exists in our knowledge of the human genome due to the unknown function of the uncharacterized proteome.4 This gap, the dark matter of the human genome encompassing the non coding RNAs and uncharacterized protein coding sequences constitute over half of the known genes. New therapeutics and early diagnostic markers are urgently needed for major illnesses including, cancer, cardiac diseases, diabetes, infection, metabolic and mental disorders and neurodegenerative diseases.6
Reasoning that the uncharacterized proteins may offer new opportunities for therapeutic intervention, we have embarked on a systematic characterization of the Open Reading Frames (ORFs) present in the human genome.7–11 The Genetic Association Database (GAD) provided a starting point to identify disease relevant ORFs of the dark Matter proteome.12 The ORF hits from the GAD were complemented with other disease-oriented databases including the MalaCards13 and the NextBio Meta analysis tool. From these analyses, 800 ORFs were identified relevant to diverse diseases and disorders. These ORFs were characterized using a streamlined approach we have recently established.10,11,14,15 Using phenome-Genome analysis, protein motif and domains, functional annotation and mRNA and protein expression analysis tools, a detailed knowledgebase of the dark matter ORFs was established. Classes of druggable proteins (enzymes, transporters, channel proteins and receptors) and secreted biomarkers were identified. Disease association to Cancer, diabetes Type I and Type II, Systemic Lupus, Rheumatoid Arthritis, Crohn’s Disease, Leprosy, Hepatitis C, Attention Deficit Disorder, Alcoholism, Cardiac diseases, Erectile Dysfunction and for various genetic disorders was seen with the dark Matter ORFs. The database of novel ORF proteins should aid in therapeutic target discovery for cancer and other diseases.
Genome analysis
The genome analysis was performed using the Genetic Association Disease, GAD database,12,16 the University of California Santa Cruz, UCSC Genome Browser,17 the ensembl Genome Browser,18 the National Center for Biotechnology Information, NCBI Gene, NCBI Aceview,19 the Sanger Institute Catalogue Of Somatic Mutations In Cancer, COSMIC,20 the Integrated Drug Discovery platform, canSar v2,21 cBioPortal,22 International Cancer Genome Consortium, ICGC,23 the Roche Cancer Genome Database, Mutome DB,24 Expression Quantitative loci browser, eQTL,25 Genotype –Tissue Expression Project, GTEx26 and the European Bioinformatics Institute- European Molecular Biology Laboratory, EBI-EMBL tools.
Transcriptome analysis
The transcriptome analysis of the uncharacterized proteins was undertaken using the NCBI-UniGene, SAGE Digital Anatomical Viewer,27 Cancer Genome Anatomy Project, CGAP,28 Oncomine microarray analysis tool,29 the Array express,30 The Gene Expression Omnibus, GEO and the Gene Indices from the Dana Farber Cancer Institute.31
Proteome analysis
The uncharacterized ORFs were analyzed for structural proteomics using the UniProt Knowledge base, UniProtKB,32 Swiss Expasy server, Protein Database, PDB, post translational modification sites at Expasy,33 PredictProtein,34 MEta Sever for Sequence Analysis, MESSA35 and the I -TASSER server.36 The protein motifs and domains were analyzed using the NCBI Conserved domain database, CDD,37 The PFAM,38 ProDom,39 InterProscan4,40 HMMER,41 Signal P server,42 and the Eukaryotic Linear Motif prediction, ELM.43 The protein expression analysis was performed using multiple tools including the Human Protein Atlas, HPA,44 the Multi Omics Proteins Expression Database, MOPED,45 the Human Protopedia Reference Database, HPRD,46 the Human Proteome Map47,48 and Proteomes database, proteomics dB.49
Knowledge-based Data mining
The knowledge databases used in the study included GeneCards,50 GeneAtlasm,51 NextBio meta analysis tool,52 The MalaCards,13 On line Mendelian Inheritance in Man, OMIM, Human Genome Nomenclature Committee, HGNC,36 Gene Ontology, amigo,53 NCBI SNP database, the NCBI Phenome-Genome integrator, PheGenI,25 the Expression Quantitative Trait Browser, eQTL,26 the NCBI Clinical Variations database, ClinVar,54 Ensembl database, The Strings Interactome,55 BioGrid,56 IntAct,57 the KEGG pathway and the DAVID functional annotation tool.58
Data analysis
The entire database of GAD, Human Protein Atlas, HPA and UniGene was downloaded as comma separated value (csv) files and the Excel filtering tool was used to scan for the ORFs. Batch analysis of the ORF database was performed for canSar, the MOPED, the DAVID annotation tool, the Human Proteome Map, Proteomics dB, the HPRD, the PheGenI and the eQTL browser. The NextBio Meta Analysis was performed for the individual ORFs.
For the bioinformatics tools, the basic options were used as default. The PheGenI tool was used with P-values set at <10-5 and the R-Squared at 0.3.
Big data verification was performed by verification using two independent experiments. Prior to using a bioinformatics tool, a series of control query sequences were tested to evaluate the predicted outcome of the results.10,11,15
Landscape Complexity of the Dark Matter of the Human Genome
The current landscape of the human genome in the context of coding and non-coding sequences is shown in Figure 1. The dark matter of the human genome encompassing the non coding RNAs (ncRNAs), the uncharacterized proteins and Open Reading Frames (ORFs) currently account for over half of the HGNC approved protein coding sequences (10,225/19,052). In attempt to establish disease relevance for the protein ORFs, multiple disease-oriented databases (Genetic Association database, The MalaCards from Gene Cards and the NextBio Meta analysis tool) were used. The GAD was enriched for positive genetic association evidence and 2,375 genetic polymorphisms associated with ORFs were identified. These ORFs were filtered for disease classes using three filters i) broad phenotypes, ii) disease classes and iii) Medical Subject Headings terms. The Dark matter ORFs-related polymorphisms were detected for diverse diseases.
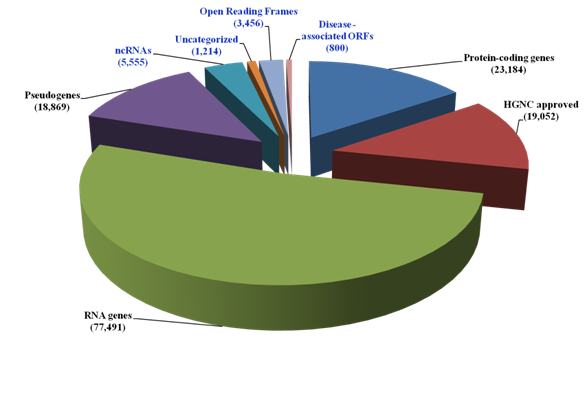
Figure 1 The human genome Dark Matter landscape
The current number of genes (known and uncharacterized) was obtained from GeneCards, HGNC and UniProt. The disease associated ORFs were identified from the Genetic Association database, the MalaCards and the NextBio Meta analysis tool. The dark matter of the genome is shown in blue.
From these analyses, 800 ORFs were identified (Supplemental Table, S1). These ORFs termed as Dark Matter ORFs, were verified by batch analysis using the GeneALaCart tool from the GeneCards. From these studies a comprehensive knowledgebase (Dark Matter ORFs) was created (Supplemental Table, S2). Additional verification of the ORFs came from HGNC, UniProt and UniGene databases. This Dark Matter ORFs were classified into non coding RNAs (n=47), uncharacterized (n=591) and known gene –related (n=164). Among these 800 ORFs, 415 ORFs were positive for genetic association evidence in the GAD.
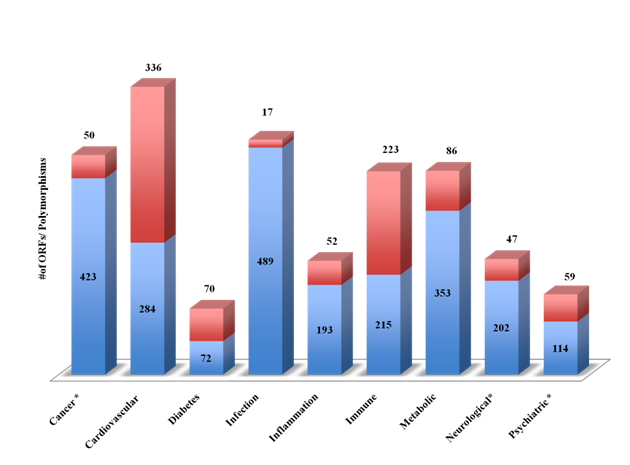
Using advanced filtering option, the Dark Matter ORF database of genes was classified into broad classes of therapeutic areas (Figure 2). The number of ORFs for each therapeutic areas (blue bars) and the number of ORFs with genetically associated SNPs (red bars) is shown in Figure 2. Largest number of ORFs was associated with three major classes: infection (n=490), cancer (n=424) and metabolic disorder (n=352). Association of the Dark Matter ORFs with multiple diseases encouraged us to undertake a further detailed analysis to provide an atlas of novel genes for target discovery.

Figure 2 The Dark Matter ORFs in therapeutic areas
The Dark Matter ORFs were identified from the Genetic Association Database (GAD), the MalaCards and the NextBio Meta Analysis tool. The disease-associated ORFs (protein coding and ncRNAs) shown in figure 1 were categorized into broad classes (blue) and genetically associated ORFs (red).
Gene ontology
The 800 Dark matter ORFs were batch analyzed for Gene Ontology (GO) using the DAVID functional Annotation tool. Additional GO verification was obtained from the GeneALacart, the GeneCards, the canSar and UniProtKB databases (Figure 3). The ORFs were classified according to the localization (panel A), function (panel B) and process (panel C). Druggable targets (enzymes, n=92, integral membrane, n=237, transporters, n=19, receptors, n=6) and putative secreted ORFs (extracellular region, n=82) were identified by the GO analyses. Thirty-one of these extracellular ORF proteins harbored classical signal peptide sequence at the N-terminus. Further, the cell localization data (see panel A) indicated various sub-cellular compartments including the Cytoplasm, Nucleus, Golgi, Mitochondria and Vesicles. The ORFs were classified into distinct binding classes including carbohydrate, lipid, metals, nucleotide and proteins (Panel B). The Dark Matter ORFs were also associated with diverse processes/pathways including apoptosis, cell cycle, differentiation, DNA repair, metabolic, signal transduction etc. (Panel C). These results provided additional insight into the nature and putative function of the Dark Matter ORFs and suggested their involvement in multiple cellular processes involving growth control and regulation.

Figure 3 Gene Ontology of the Dark Matter ORFs
Putative classes of the Dark Matter ORFs inferred from Gene Ontology tools (DAVID, GeneCards and UniProtKB). Panel A) localization; Panel B), function and Panel C) process.
Gene expression
The availability of numerous gene expression analysis tools for both mRNA and protein allowed us to establish the gene expression profile of the dark Matter ORFs in normal and diseased tissues. The mRNA expression in normal tissues was analyzed using the UniGene (Expressed Sequence Tag, EST) and the NextBio Meta Analysis tool (Microarray datasets). Additional verification of mRNA expression for normal and tumor tissues was derived from the Oncomine Microarray database. Multiple protein expression analysis tools (MOPED, HPRD, HPA, Human Proteome Map, and the Proteomics dB) were used to verify the protein ORF expression in diverse normal tissues including body fluids and tumor tissues (Supplemental Table, S3).
ORF expression (mRNA and protein) was detected in diverse body fluids including blood, bone marrow, bronchoalveolar lavage, cerebrospinal fluid, cerumen, ascites, milk, proximal fluid, tear, saliva, semen, synovial fluid and urine (n=102/800). A total of fifty-eight ORFs were found to have a classical signal peptide sequence at the N-terminus; eleven of these signal peptide-containing ORFs were detected in the body fluids. Further, tissue- and developmentally regulated expression was seen for diverse ORFs (Figure 4). In the adult, largest number of ORFs expression was seen in the testis (n=319) followed by the brain (n=135). Fetal, infant and adult-restricted expression was also seen for the ORFs. A correlation of expression at the mRNA and protein level across multiple datasets was seen in approximately10% of the ORFs (87/800).
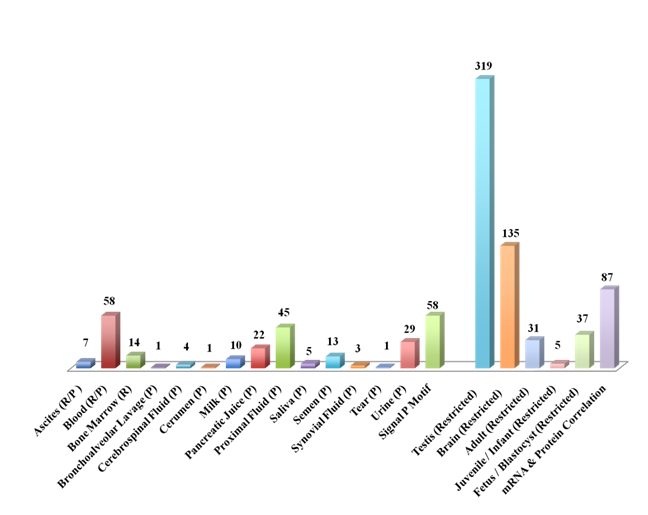
An ORF, C19orf10 interleukin 27 working designation,59,60 present in the pancreatic juice, bronchoalveolar lavage, milk, semen and urine was associated with pancreatic, prostate and hepatic carcinomas. This ORF is also associated with other diseases including Asthma, Cardiac transplant rejection, Pancreatitis, Encephalomyelitis, HIV-1, Genetic Predisposition to Inflammatory Bowel Disease and Crohn's disease.61 This novel putative cytokine ORF has potential for diagnostic marker development. Other ORFs present in the pancreatic juice included c19orf173, C14orf10, C5orf52, C12orf56 and C19orf43 and may offer target potential for pancreatic diseases including cancer.
The urinary ORFs included C11 orf52, C17orf37, C19orf10, C2orf54 and C9orf142. The C17orf37, a selenoprotein, located at the ERBB2 locus is associated with Breast cancer susceptibility62,63 and may provide a non-invasive urinary detection. The milk ORFs encompassed C6orf15, C11orf52, C11orf67, C16orf178, C19orf10 and C21orf23.
Further, the detection of the Dark Matter ORFs in other body fluids including the proximal and synovial fluids, semen and cerumen opens a new potential for minimally invasive and noninvasive diagnosis.
In addition to protein coding ORFs, ncRNAs were also detected in the body fluids. This included bone marrow (C21orf82, C21orf93,C21orf94, C21orf96) and tear (C9orf27).
The presence of protein coding ORFs and the ncRNAs in diverse body fluids offers a rationale for non-invasive methods of detection in multiple therapeutic areas.
Phenome-genome analysis of the dark matter ORFs
The Phenotype-Genotype Integrator (PheGenI), a phenotype-oriented resource facilitates follow-up studies from GWAS and allows prioritization of variants. An expression quantitative trait locus (eQTL) represents a gene marker (locus) in which variation between individuals is associated with a quantitative mRNA gene expression trait.64 The eQTL results encompass 1) a SNP marker; 2) the gene expression levels; and 3) a measure of the statistical association between the two in a study population, such as the P-value.
The Phenotype-Genotype Integrator (PheGenI) was used to establish phenotypic traits associated with the ORFs. Genetically associated traits were seen for 227/800 ORFs in diverse therapeutic areas (Figure 5). We next performed eQTL analysis for the phenotype positive ORFs. The ORFs were further analyzed using the eQTL browser for eQTL traits. Genotypes for the OncoORFs were selected for exons, introns, near gene and Untranslated Region (UTR). From the output of results, disease traits were enriched (Supplemental Table, S4).
Twenty-nine ORFs were identified for eQTL traits in major diseases and disorders including
Association with diverse traits was seen for some of the ORFs: C2orf43 (prostate cancer, Cholesterol, Echocardiography, Aspartate Aminotransferases); C11orf10 (Crohn’s Disease, Cholesterol, Phospholipids, Lipoproteins); C8orf13 (Systemic Lupus, Rheumatoid Arthritis) and C6orf204 (Renal Carcinoma, Electrocardiography).
On the other hand, unique association was also seen with some of the ORFs in Non Small Cell lung carcinoma (C3orf21); prostate neoplasm (C18orf34); Leprosy (C7orf44); Chronic Hepatitis C (C20orf194); Blood pressure (C14orf118); Alcoholism (C3orf59); Attention Deficit Disorder, (C12orf28); Psychomotor Performance (C11orf73) and Erectile Dysfunction (C9orf3).
Clinical variations
We next established the clinical relevance of the ORFs using the NCBI Clinical variations database, ClinVar54 for pathogenicity and presence in clinical samples. Twenty-one ORFs were identified with clinical correlation and pathogenicity (Supplemental Table, S5).
Clinical significance was seen in developmental delay (C2orf43, C2orf48, C2orf50, C2orf61, C2orf73, C2orf91, C5orf42, C15orf41, C17orf105); Retinitis pigmentosa (C2orf71, C8orf37); Melanoma (C2orf16, C5orf42, C12orf57, C17orf104) and non small cell lung cancer (C10orf11, C17orf31); Oculocutaneous albinism (C10orf11); Recessive hypo mineralized amelogenesis imperfect (C4orf26); Amyotrophic lateral sclerosis and/or frontotemporal dementia (C9orf72); Temtamy syndrome (C12orf57); Spastic paraplegia 55, autosomal recessive (C12orf65); Chiari malformation type II (C17orf105); Autosomal dominant progressive external ophthalmoplegia (C10orf2); Spastic paraplegia 55, autosomal recessive (C12orf65); Anemia, Congential Dyserthropoitic (C15orf41) and Neurodegeneration with brain iron accumulation 4 (C19orf12). Dark Matter ORFs in multiple diseases.
In an attempt to develop a therapeutic area –related ORFs for druggableness and diagnostic markers identification, we next investigated the Dark Matter database for SNPs associated with major diseases. Figure 5 shows the number of dark matter ORFs and the SNPs associated with major therapeutic areas (cancer, cardiovascular, diabetes, infections, inflammation, immune, metabolic, neurological and psychiatry). About half of the ORFs were associated with infectious diseases and cancer (n=490, 424/800 respectively). Cardiovascular and immune disorders were associated with the largest number of SNPs (n=338 and 225 respectively). Clinically relevant SNPs were identified using the NCBI ClinVar and were found in cancer, neurological and psychiatric illnesses.
Figure 6 shows the complex landscape of the dark matter ORFs across multiple diseases. A considerable overlap with multiple diseases was seen for most of the ORFs in the database. About 6% (56/800) of the ORFs showed a high degree of selectivity to distinct therapeutic areas. For example, enrichment was seen for cancer (C7orf71), infections (C1orf 185, C1orf198, C7orf70, C9orf114, C11orf84, C19orf47), immune (C6orf100, C10orf64), inflammation (CXorf22) and neurological disorders (C20orf7).
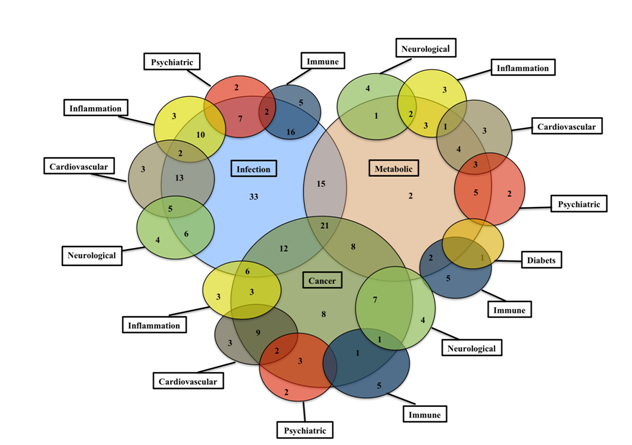
The C7orf71, a testis-enriched novel ORF showed strong association with neuroendocrine, brain, gastric tumors, lymphomas and HIV-1.65 The C19orf47, an Influenza viral infection-associated ORF, is a putative secreted protein with signal peptide sequence at the n-terminus and is present in blood plasma. This ORF offers a less invasive diagnostic potential. The C6orf100 is associated with Graves' disease, Systemic Lupus and Autoimmune endocrine diseases.66,67 The C10orf64, a WD repeat- and FYVE domain-containing protein 4 is associated with Systemic lupus, Autoimmune skin disease, Rheumatoid arthritis and Chemdependancy.66,68 The C20orf7, a probable mitochondrial methyltransferase| NADH dehydrogenase [ubiquinone] 1 alpha sub complex assembly factor 5, is associated with Paralytic syndrome, Mitochondrial Complex I Deficiency, Chronic fatigue syndrome and Chemdependancy.69–71
These results underscore the overlapping involvement of the ORFs in multiple diseases and emphasized the ORF landscape complexity.
Cancer mutations of the dark matter ORFs
The Catalogue of somatic mutations in cancer database, COSMIC has comprehensive mutation data for both the known and uncharacterized proteins.17 The entire COSMIC database was downloaded and enriched for the dark matter ORFs (Supplemental Table, S6).
The ORFs harbored diverse mutations (nonsense, missense, deletions, insertions, frame shifts, in-frames, homozygous and heterozygous) as shown in Figure 7. Heterozygous mutations accounted for the largest number of mutations (81.57%) followed by substitution missense mutations (12.5%). Major COSMIC mutations were present in Breast (n=1,874), endometrium (n=6,528), kidney (n=1192), Large Intestine (n=7,962), liver (1,225), lung (n=7,445), esophagus (n=813), Ovary (n=833), pancreas (n=323), prostate (n=569), skin (n=1,342), stomach (n=340) and urinary track (n=861).
Motif and domains analysis

To develop further insight into the nature of the Dark matter proteins, the ORFs were analyzed for protein motif and domains. The GeneALaCart tool was used to batch analyze the ORFs for the InterPro/UniProt Domains and Families.72 In addition, the NCBI Conserved Domain Database, CDD,37 the Protein Family, PFAM38,73 the Biosequence analysis using profile hidden Markov models, HMMER,41 the Protein Domain Analysis, ProDom,39 UCSC Genome Browser74 and SignalP42 bioinformatics tools were used to analyze the Dark Matter ORFs. The post-translational modification sites, binary interactions and protein architecture and complexes data were obtained from the HPRD database batch analysis (Supplemental Table, S7).
Noncoding RNAs in diseases
Forty-seven ncRNAs were identified among the Dark Matter ORF database. These encompassed long intergenic RNAs, Linc RNAs (n=23), antisense transcripts/Linc RNAs (n=31)and pseudogenes (n=4).These ncRNAs showed genetic association with diverse diseases including cancer, cardiovascular, diabetes, infections, inflammation, metabolic, neurological and psychiatric illnesses. Distinct ncRNAs were associated with type I diabetes (C6orf208) and type 2 (C6orf27, C14orf70). The ORF, C1orf133|SERTA domain containing 4 antisense RNA 1, a putative uncharacterized protein showed association with celiac disease. The C18orf16|Aquaporin Isoform Delta 4 Antisense RNA was associated with both type 1 and type 2 diabetes. The C9orf29| leucine rich repeat containing 37, member A5, pseudogene showed association with bladder and prostate carcinomas, multiple myeloma and plasmacytoma, Irritable bowel syndrome and Cytomegalovirus infection. The C14orf55 | ADAM metallopeptidase domain 20 pseudogene 1 was associated with Multiple myeloma/plasmacytoma, Cancer of head and neck, Prostate cancer and Coronary arteriosclerosis. The C20orf191|nuclear receptor corepressor 1 pseudogene 1 was associated with Systemic lupus erythematous, Systemic inflammatory response syndrome, Lipoproteins, HDL and Uveitis. These results provide novel ncRNA targets for diverse diseases.
From these analyses, the Dark Matter ORFs were annotated into functional classes of proteins (Table 1). Protein families including antigens, carrier proteins, enzymes, nucleotide/metal binding, receptors, mitochondrial chaperones, phosphoproteins, secreted glycoproteins, selenoproteins, transporter/sorting proteins, vacuolar proteins and Zinc finger proteins were identified among the Dark matter ORF proteins.
Protein family |
Number of dark matter ORFs |
Antigen |
8 |
Antisense |
35 |
Carbohydrate/heparin binding |
6 |
Channel |
4 |
Enzymes |
118 |
Glycoproteins |
41 |
Metal binding |
24 |
Nucleotide binding |
55 |
Phosphoprotein |
18 |
Protein Binding |
52 |
Receptors |
20 |
Secreted |
57 |
Transcription factors |
32 |
Transmembrane |
122 |
Transporters |
9 |
Vesicular |
1 |
Table 1 Protein characterization of the Dark Matter ORFs: Motifs and Domains
We have embarked on systematically deciphering the uncharacterized ORFs, the Dark Matter proteome in the human genome. These ORFs are the least analyzed putative proteins in the genome and continue to remain a mystery. Realizing a target potential for these novel proteins for druggableness and diagnostic markers development, a comprehensive analysis of the ORFs was undertaken. Omics characterization was performed for 800 disease-oriented ORFs in the human genome. An atlas of the Dark Matter proteome was generated encompassing details on the somatic mutations, baseline mRNA and protein expression, protein motif and domains, disease traits and clinical relevance. Over half of the ORFs (416/800), analyzed in the study showed positive genetic association with diverse diseases and disorders. The disease landscape complexity of the ORFs offers an advantage to develop a single ORF target for novel therapy and diagnosis for diverse diseases. Omics correlation.
In a recent study, Zhang et al.,75 performed an extensive proteogenomic characterization of colon and rectal tumors using The Cancer Gene Atlas (TCGA) samples. Their results indicate a lack of correlation of gene amplification, mRNA and protein expression for over two thirds of the samples analyzed. Only a small number of genes showed a strong correlation of gene expression at the mRNA versus protein levels. This is not surprising in view of the issues such as stability and post -translational modification and regulation of protein expression.76 Our results concur with these findings: 87/800 ORFs (about 10%) showed correlation of expression at the mRNA and protein levels. For druggable target prediction as well as pathway mapping, such a correlation can facilitate target prioritization. Our study predicts secreted proteins as measured by mRNA and protein expression in diverse body fluids (102/800 ORFs) including blood, cerebrospinal fluid, synovial fluid, plasma, serum, bone marrow, Bronchoalveolar lavage, ascites, milk, pancreatic juice, semen, saliva, tear and Urine. Signal peptide sequence was detected in 58/800 ORFs. The baseline expression of the Dark Matter ORFs in the body fluids provides rationale for further investigation into secreted biomarkers characterization.
The Noncoding RNAs
The ncRNA class encompasses diverse non coding RNAs such as the long intergenic RNAs, micro RNAs, Small nuclear ribonucleic acids, Small interfering RNAs, the antisense transcripts and the pseudogenes. The ncRNAs play key roles in the regulation of gene expression at the transcriptome and proteome levels. Long considered as the Dark matter of the genome, these ncRNAs are increasingly becoming important in diagnosis and therapy of diverse diseases.77,78 The forty-seven ncRNAs comprising linc RNAs, antisense RNAs and pseudogenes discovered in this study, offer new target potential for diverse diseases including autoimmune disease, cancer, cardiovascular and inflammation. Further, the detection of some of the ncRNAs in the body fluids such as the bone marrow and tear offers diagnostic potential.
Genome to Phenome association
Disease-associated phenotypes were seen by eQTL analysis for 29 ORFs in diverse diseases including autoimmune diseases, cardiovascular diseases, cancer, infections, inflammation, and psychiatric disorders. Association with multiple traits was seen for some of the ORFs, for example,C2 or f43 (prostate cancer, Cholesterol, Echocardiography, Aspartate Aminotransferases); C11orf10 (Crohn’s Disease, Cholesterol, Phospholipids, Lipoproteins); C8orf13 (Systemic Lupus, Rheumatoid Arthritis) and C6orf204 (Renal Carcinoma, Electrocardiography). On the other hand, unique phenotypic association was also seen with some of the ORFs such as in Non Small Cell lung carcinoma (C3orf21); prostate neoplasm (C18orf34); Leprosy (C7orf44); Chronic Hepatitis C (C20orf194); Blood pressure (C14orf118); Alcoholism (C3orf59); Attention Deficit Disorder (C12orf28); Psychomotor Performance (C11orf73) and Erectile Dysfunction (C9orf3). These ORFs may offer novel opportunities for diagnosis and therapy of various diseases.
Clinical relevance
The NCBI ClinVar tool identified 21 ORFs in the dark matter ORFs, which are highly clinically relevant in diseases of the eyes and skin, inherited genetic disorders, developmental disorders and the neurodegenerative diseases. For example,
Truncating mutations in C2ORF71 were identified in three unrelated families causing autosomal-recessive retinitis pigmentosa (RP). Clinically, patients with mutations in C2ORF71 show signs of typical RP.79 The C2orf71 mutations were also associated with developmental delay.80 The C2orf71 encodes a transmembrane photoreceptor. Another ORF, the C8orf37| small talk protein is also highly correlated with RP and Cone-rod dystrophy (CRD). These authors showed by Immunohistochemical studies that the C8orf37 protein is localized at the base of the primary cilium of human retinal pigment epithelium cells and at the base of connecting cilia of mouse photoreceptors.81 The C8orf37 protein encodes a ciliary photoreceptor. Homozygous truncating mutation and missense mutation in C12orf57 | likely ortholog of mouse gene rich cluster was shown to be involved in the pathogenesis of a clinically distinct autosomal-recessive syndrome form of colobomatous microphthalmia, a developmental eye disorder.82 The C12orf57 protein encodes a novel uncharacterized protein.
The C4orf26 loss of function mutation was associated with of recessive hypo mineralized amelogenesis imperfecta (AI). This is a disease in which the formation of tooth enamel fails.83 The C4orf26 encodes a putative extracellular matrix acidic phosphoprotein expressed in the enamel organ. Joubert syndrome (JBTS) is an autosomal-recessive disorder characterized by a distinctive mid-hindbrain malformation, developmental delay with hypotonia, ocular-motor apraxia, and breathing abnormalities. Rare compound-heterozygous mutations in C5orf42 have been shown to cause JBTS in French Canadian individuals.84 The C5orf42 encodes a novel uncharacterized, transmembrane coiled-coiled protein.
An expansion of a non coding GGGGCC hexanucleotide repeat in the gene C9orf72 was shown to be strongly associated with autosomal-dominant frontotemporal dementia (FTD) and amyotrophic lateral sclerosis (ALS), genetically linked to chromosome 9p21.85,86 The gene C9orf72 encodes for a GDP/GTP exchange factor (GEF). Mutations in C10orf11 | Fasting-induced gene protein, a melanocyte-differentiation gene, has been shown to cause autosomal-recessive albinism, which is a hypopigmentation disorder.87 The C10orf11 encodes for a Leucine-rich repeat, small nuclear Ribonucleoprotein A. Missense mutations in the C15orf41 gene were implicated in congenital dyserythropoietic anemias.88 The C15orf41 protein with two predicted helix-turn-helix domains encodes a novel restriction endonuclease, bearing the Holliday junction resolvase family signature. An 11bp deletion in the C19orf12 | spastic paraplegia 43 (autosomal recessive) has been shown to cause neurodegeneration with brain iron accumulation.89 This gene encodes a Mitochondrial Transmembrane Iron binding protein.
These results demonstrate that the uncharacterized components of the human genome provide valuable clues to understanding biology, genetics of diseases, novel diagnostic and therapeutic opportunities.
Results presented in this study shed light on the uncharacterized ORFs, the Dark Matter of the human proteome. Verification of motifs and domains, expression in body fluids and association with diverse disease phenotypes offer novel therapeutic and biomarkers potential for these ORFs. Protein classes encompassing channel proteins, enzymes, receptors, secreted molecules, transcription factors and transporters were identified. The atlas of the 800 novel proteins developed in this study provides an attractive starting point for accelerated drug discovery and diagnostic markers development for major diseases.
RN was responsible for conceiving, data mining, batch analysis and overall execution of the project. Data validation and visualization was performed by APD. MJC was responsible for Motifs and domains analysis. PB was responsible for mining the COSMIC and the NextBio databases. SH was responsible for mining the HPA, Roche Mutome and HPRD databases.
We thank Dr. Stein, GeneCards team for generous permission to use the GeneALaCart tool, Dr. Libby Montague, Kolker Laboratory, MOPED Team for the batch analysis of the ORFs; the NextBio Meta analysis tool for permission to use the dataset; the CanSar, cBioPortal, the TCGA and COSMIC for the mutations dataset, the Human Protein Atlas, Human Protein Reference Database and DAVID functional annotations tools for various datasets. We especially thank the human proteome map and the proteomes dB (Dr. Mathias Wilhelm) for the protein expression datasets. This work was supported in part by the Genomics of Cancer Fund, Florida Atlantic University Foundation. We thank Jeanine Narayanan for editorial assistance.
The author declares no conflict of interest.

©2015 Delgado, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.