MOJ
eISSN: 2374-6920


Review Article Volume 4 Issue 5
Sage-N Research, Inc., Milpitas, California, USA
Correspondence: David Chiang, Sage-N Research, Inc., Milpitas, California, USA
Received: July 10, 2016 | Published: November 30, 2016
Citation: Chiang D. How to identify low-abundance modified peptides with proteomics mass spectrometry. MOJ Proteomics Bioinform. 2016;4(5):298-304. DOI: 10.15406/mojpb.2016.04.00133
Characterization of low-abundance, modified peptides (LAMPs) is uniquely valuable for research in early detection of cancer and infections, and indeed for all molecular biology research. However it challenges proteomics mass spectrometry due to low signal-to-noise. With parts-per-million mass accuracy for both intact (“precursor”) and fragment ion masses available from diverse mass spectrometers, it becomes feasible but requires capable analytics-i.e. mathematical and computing techniques to infer insights from patterns hidden within the data. This is best done with simple mathematical transformations of raw mass data applied at a large scale, which ensures subtle data patterns arise from the data and not processing artifacts. Patterns should be visually clear to the human eye, which is still the very best data analysis tool.
Traditional analytics rely largely on search engine “similarity” scores (e.g. XCorr, ion-score, hyperscore) that quantify the degree of similarity between the measured spectrum and a predicted spectrum, the latter derived from a candidate peptide sequence using a sequence-to-spectrum ionization model. They evolved mainly for abundant peptides with robust spectral signals. Efforts to improve the search engine score, for example by incorporating accurate fragment data, have had limited success or worse. Some ad hoc efforts identify unexpectedly numerous peptides having near-identical mass, charge, and chromatographic retention time - i.e. same isotopic envelopes at the same split-second in a multi-hour experiment - better explained by faulty analytics.
Current workflows have two intractable problems for LAMPs. First, similarity scores have a dependence on charge and peptide length that introduces complex artifacts impossible to reverse. Second, mass accuracy of fragment ions has been mainly used pre-search to narrow the search space rather than as a post-search filter, which worsens noise susceptibility (Figure 1).
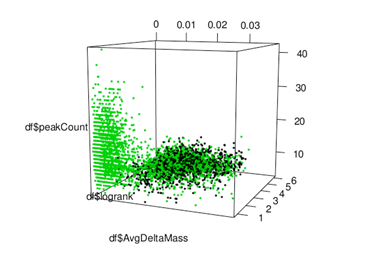
Figure 1 Sorcerer Score 3D data cube with S-scoreʼs three components, showing clear separation between likely-correct (green) and likely-incorrect (green & black) candidate peptides [non-decoys in green; decoys in black].
On one hand, this reduces obviously-incorrect, out-of-fragment-mass-range candidates, which sounds good on the surface. Counter intuitively, it increases noise susceptibility because incorrect peptides that randomly get through would be indistinguishable from correct ones with respect to fragment mass. In other words, fragment mass, one of the most fundamental information from tandem mass spectrometry, is rendered useless as a correct-vs-incorrect discriminator. Instead, the robust way is to allow enough out-of-fragment-mass-range candidates to define the background, and to use fragment mass as a post-search filter to separate outliers from such background.
A cross-correlation similarity score has the best noise-suppression among popular search score types, all of which are expressible as a vector dot-product. It is therefore the best type of search engine score for LAMPs. However, for the post-search filter, the score rank is a statistically cleaner surrogate with good correct-vs-incorrect discriminating power. This is akin to a college recruiting on the basis of rank (i.e. valedictorians) vs. the GPA score.
We present a new simplicity-focused analytics methodology, called Sorcerer Score(tm) that allows LAMPs to be rigorously identified within a hypothesis-driven framework based on high-accuracy precursor and fragment mass data. Accurate peptide ID is fundamental to accurate protein quantitation and post-translational modification (PTM) analysis.
The general approach is to: (1) use standard “target-decoy” cross-correlation search with a wide mass range and “implicit decoy” sequences (i.e. known-wrongs within the target search space), (2) pre-filter out peptides with excessive mass error and implicit decoys, and then (3) compute the following discriminant score:
S-score=peak Count/p0- Avg Delta Mass-log (Rank+1)/r0.
The most important component is Avg Delta Mass, which is the positive weighted average of precursor mass error (adjusted for in-measurement mass changes and calibration skew) and the average fragment mass RMS error. Peak Count is the number of matched fragment ions between measured and predicted spectra. The S-score is compatible with any tandem mass spectrometer with sufficient mass accuracy.
S-score has units of mass (amu) and is derived primarily from raw mass data. It has a high value when the average mass error is small, many fragment ions are matched, and the similarity score rank is high, which are most of the key information from tandem mass spectrometry. Clearly it is a correct-vs-incorrect discriminator in a qualitative sense. What is surprising is that such a simple figure-of-merit can be quantitatively powerful, which we illustrate with data.
To the chemistry-inclined, Sorcerer Score may be viewed as “digital chromatography” whereby a “mixture” of candidate peptide IDs are multi-dimensionally enriched using the parameters that comprise the S-score. Decoys in the search space act as a solvent that preferentially flushes out incorrect IDs to leave enriched likely-correct IDs concentrated among the top ranks. This is the data-mining tradeoff: more solvent (i.e. decoys through CPU power) for more purity (i.e. lower FDR). Conversely, weak “demo-quality” analytics that minimize CPU time, using over-optimistic algorithms that underestimate error rates and confuse noise with signal, is likely the main cause of irreproducibility in high-mass-accuracy proteomics.
Just as chemical separation would use different chemicals matched to the sample, peptide identification can use additional filtering parameters matched to the dataset and the biological system. For example, certain PTMs yield characteristic fragment ions useful for further filtering. This is why an open data-mining “platform”, as opposed to a closed push-button “program”, is needed for high-value proteomics analysis - the only kind that can sustainably justify pricey mass spectrometers.
A human expert aided by a computing platform is the norm for diverse data-driven fields like stock market analysis, weather-forecasting, credit card fraud detection, etc. Unlike computers, human experts can use unanticipated meta-information to narrow down choices in deep analysis, like Sherlock Holmesʼ dog that didn’t bark. Clearly, it is impossible to code a computer to check for all the ancillary things that do and do not happen.
Per the 80/20 Rule, we expect that any and all fully-automated workflows can only identify the easier 80%, with the rest requiring expert data-mining for ever deeper analysis. Every experiment will have a different cost-benefit tradeoff. High-value experiments will benefit from data-analysis-as-a-service from experienced proteomic data scientists.
The Sorcerer Score concept is simple by design but challenging in practice due to the computation. A typical 100K-spectra dataset cross-correlation searched with multiple PTMs and extensive decoys, while keeping top-200 candidate peptides, results in 20million candidate peptides from which ~15K to 20K correct IDs are extracted. A 1M-spectra dataset would have 200M candidate peptides. High-performance multi-million data point analysis requires professional server-class development that goes beyond writing a C program on a PC.
When the 3 components of S-score are plotted in a 3-D cube, a few thousand non-decoy points (including duplicates with high sequence homology) become clearly separated from the background. (See Figure 1 above) The S-score of any point is its distance to the “S-score=0” plane along the Avg Delta Mass axis.
The parameters (p0, r0) define the tilt of the separation plane (S-score=X) in the 3-D cube. The cutoff threshold ʻXʼ can be set to trade off ID quality vs. estimated FDR. Notably, we found the results to be robust. For example, to achieve FDR ~1% in our sample dataset that has ~2K correct peptides, basically the same ~20 decoys are responsible for the FDR. These ~20 decoys are relatively independent of p0 and r0. We believe robustness translates to research reproducibility.
The core Sorcerer Score analytics is implemented in a short R script within the SX1301 MUSE script which can be modified or customized by the user or by Sage-N Research. All the plots shown were interactively created in R off-line on a Mac by re-running the R part of SX1301 on data files generated on a SORCERER system. Most of them are also auto-generated in a PDF file by the SX script that may be viewed online within SORCERER.
We believe Sorcerer Score uniquely enables deep proteomics by presenting peptide IDs close to their raw data from using simple-to-understand analytics. Its foundation is based on well-established components, namely the cross-correlation search engine (John Yates Lab, Scripps), target-decoy search (Steve Gygi Lab, Harvard), and a rigorous peptide-to-protein framework (Ruedi Aebersold Lab, ETH Zurich).
Unlike opaque software that report peptide IDs not readily verifiable, Sorcerer Score respects the integrity of the science by being transparent and hypothesis-driven, and by presenting data-driven evidence that can be drilled down to any level by scientists.
An anonymous phospho-peptide dataset (~6K spectra) from a Thermo Q-Exactive mass spectrometer is searched and then automatically processed using the SX1301 GEMYNI script on a SORCERER system. The spectra comprise high-accuracy b-/y- fragment ions (ʻHCDʼ).
The search conditions are as follows on SORCERER (masses are rounded to nearest amu for illustration):
These conditions allow for a single labile phosphorylation site (precursor dMass ~+98amu). Since this is a CID-only dataset (high-accuracy ʻbʼ and ʻyʼ fragment ions), ETD terminus modifications are used to generate implicit decoys only.
The ~0.5 hour search results in ~2.8M candidate peptide IDs. After pre-filtering, the S-score is calculated for each remaining candidate ID.
For peptide IDs with ʻNʼ instances of labile phosphorylation of serine/threonine, the effective delta-mass is adjusted down by N*(-98) to account for the net -98amu (= 80+18) mass loss during collision.
For each spectrum, the non-decoy candidate peptide with the highest S-score, if there is one, is chosen. Otherwise, the highest S-score of either implicit or explicit decoy is chosen. The final enriched set of candidate peptides represent about 92% of the spectra, as about 8% of the spectra yielded no candidate peptides after pre-filter. Most figures are shown with enriched set. Manual examination showed no credible multi-peptide IDs for this dataset.
When we set the overall FDR cutoff ~1%, we identified both abundant peptides (defined as rank=1) with FDR<0.2% (1682 non-decoys+3 decoys) and low-abundance peptides (defined as rank >1) with FDR<4% (544 non-decoys+19 decoys). These include peptides with very low XCorr rank (below 200th) and with difficult-to-search phosphorylated serine/threonine. Even for abundant peptides, Sorcerer Score increases the accuracy of their analysis.
The scientific method requires hypotheses to be tested against data. Since mass spectrometry yields only mass data, such hypotheses must be mass-testable. Therefore, the search engine is best viewed as an automated hypothesis-generator and its output of candidate peptide IDs for each spectrum as independent hypotheses in the form “Peptide X generated Spectrum Y”. These hypotheses are then tested against precursor and fragment mass data. They are rejected if any observed delta-mass is excessive and conditionally accepted otherwise (Figure 2).
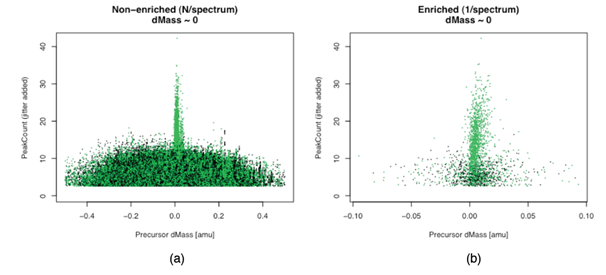
Figure 2 Scatter plot of Peak Count vs. Precursor Delta-Mass with dMass~0 [non-decoys in green; decoys in black].
For our ~6K spectra and 2.8M gross search results, we expect the small percentage of correct hypotheses to be clustered around dMass~0 with noticeably higher peak Count. Figures 2a & 2b show the plot of “peak Count vs. Precursor Delta-Mass” for both unenriched (“gross”) candidate IDs and enriched (“unique”) IDs, the latter from selecting the single best candidate peptide ID for each spectrum. Jitter is added to Peak Count for better visualization of integer values.
Notably, Figure 2a hints at the real power of target-decoy search as a high-level mass scan - a sort of ʻMS0ʼ scan, if you will - that like MS1 and MS2 scans reveal signals around certain masses. Unlike MS1 and MS2, however, it incorporates a search engine and peptide model. The signal in the ʻMS0ʼ scan is the concentration of non-decoys at certain precursor dMasses. Its utility would be clearer with a wider mass range (not shown).
Phosphorylation of serine (S), threonine (T), or tyrosine (Y) adds +80amu for the phosphate group. However, phosphorylated S or T may be unstable during mass spectrometry, resulting in beta elimination with a mass loss of 98amu (=80+18) from the phosphate group plus water. In the process, serine is converted to dehydroalanine, and threonine is converted to dehydroamino-2-butyric acid.
For example, letʼs say a serine-phosphorylated peptide enters the mass spectrometry with ʻMS1ʼ mass 1080amu (i.e. 1000amu unmodified), experiences beta elimination to become 982amu, and then fragments to produce the ʻMS2ʼ spectrum. To the search engine, this is manifested as a +98amu precursor mass error. To visually check for such peptides, we use the ʻMS0ʼ plot around dMass ~98amu.
Figures 3a & 3b show the Peak Count vs. dMass for unenriched and enriched candidate peptides IDs from the search results. They clearly show a population of likely-correct IDs with a single labile phospho-site.
From the search engine perspective, correct identification has only two requirements:
For our example, the ʻtrueʼ precursor mass is 982amu, which is manifested as a precursor delta-mass (i.e. Measured-Actual) of +98 from the ʻMS1ʼ mass of 1000. This is the mass of the pre-fragmentation ion corresponding to the spectrum.
To search for peptides with a single labile phosphorylation, the precursor mass range must include dMass ~+98amu. For double labile phosphorylation, dMass ~2*(98)amu, and so on. For example, it is possible in principle, if not in practice, to search for up to two labile phospho-sites by setting the mass tolerance to +/- 200amu.
For any variable modification search, the PTM mass is simply the mass difference between a singly modified peptide (982amu in our example) and the unmodified peptide (1000amu), or -18amu.
Conceptually, the issue is that there is no practical way to measure the mass of the ʻtrueʼ precursor that exists only inside the collision chamber for a split-second. So we have to make do with the ʻMS1ʼ mass which can be correct for stable phosphorylation, or too high by +98amu (single labile PTM), +196amu (two labile PTMs), +294amu (three labile PTMs), and so on.3,4
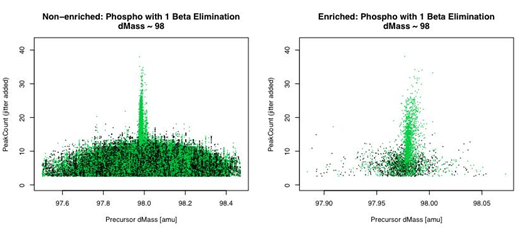
Figure 3 Scatter plot of Peak Count vs. Precursor Delta-Mass with dMass ~ 98 amu for labile phosphorylation [non-decoys in green; decoys in black].
Assuming there is no way to tell what the dMass would be from the spectrum, we have to allow for the dMass uncertainly in both the search and the post-search analysis. Essentially, we search allowing for one or more labile PTMs, then adjust the dMass for each candidate peptide based on how many -18amu labile PTMs are included, so the adjusted delta-mass (“dMassA”) is consistent with the number of labile PTMs.
For the peptide search, we must use (STY +80) and (ST -18) to search for stable and labile phosphorylation, respectively. A mass tolerance of +/- 0.5amu plus an “iso-check” set at +98amu (i.e. +/- 0.5amu for base dMassʼs of {-98, 0, +98}amu) allows for a single labile PTM.
For post-search filtering, the SX1301 script adjusts the precursor mass by N*(-98) for any candidate peptide ID containing ʻNʼ instances of (ST -18).
Since we focus on accurate deep analysis rather than formal semantics, for practical purposes we can characterize an abundant peptide as one that yields a high similarity score well-separated from other scores for a reasonably-large search space. In contrast, lower abundance peptides yield weaker signals comparable to background noise, resulting in mid to low similarity scores relative to random wrong peptides (Table 1).
LAMP type |
Accurate reported precursor mass |
Interpretable fragment ion spectrum |
Type I |
Yes |
Yes |
Type II |
No |
Yes |
Type III |
No |
No |
Table 1 Accuracy & fragment ion of LAMP
At some point, the precursor ʻMS1ʼ signal may be too small for accurate precursor mass estimation, although fragment ions may still be interpretable by a sensitive search engine. As well, a single spectrum can conceivably capture fragment ions from two or more peptides with overlapping isotopic envelopes, with its reported precursor mass incorrect for all but the dominant one.
We categorize LAMPs into three types:
In this paper we focus on Type I LAMPs only. The Sorcerer Score methodology is expected to be extensible to Type II LAMPs, albeit with less accuracy.
Figures 4a‒4d show the histogram of the original score rank of the final ID hypotheses with estimated FDR < 1%, split into {Target vs. Decoy} x {Top-10 vs. Below-Top-10}. Note that XCorr ranks to 500th are considered, but the lowest ranked target and explicit decoy are 245th and 17th, respectively.
The exponentially decreasing distribution vs. score rank is consistent with expectations of correct ID distributions.
The dramatic drop from rank 1 to 2 compared to other transitions suggest that top-ranked peptides are characteristically distinct, which supports them being considered “abundant peptides”.
To be clear, the ranks shown are the original XCorr ranks from raw search results. After final enrichment, they become the sole candidate ID hypothesis associated with each spectrum.
Avg Delta Mass is a weighted average of absolute values of the effective precursor mass error and of the average fragment mass error. The former considers any PTM adjustment and observed calibration skew. The latter is the average RMS error calculated from matched fragment ions.
The general fragment ion match is based on the Peptide Score part of the Ascore.1
For datasets where both precursor and fragment mass data are collected in the same chamber with presumably the same mass accuracy, including for our sample dataset, we can use equal weights of 0.5. This can be visually validated (see Figure 5), by ensuring a “slope=-1” line (representing the “Constant=0.5*x+0.5*y” iso-score line) reasonably fits the observed correct-vs-incorrect boundary.
Peakcount Vs. Avg delta mass
Figure 6 shows the foundation of the Sorcerer Score methodology - the clear separation between likely correct IDs with low Avg Delta Mass and high peak Count with mostly non-decoys, and a balanced decoy/non-decoy presumed incorrect IDs with the reverse. Significantly, a dividing line with a representative slope visually helps to separate most correct IDs from incorrect IDs.
Note this is a 2D projection of the 3D data-cube from Figure 1. The third dimension of log (Rank+1) helps separate correct-vs-incorrect points in the low-peak Count/high-rank region. Indeed, the correct-vs-incorrect separator is actually a tilted plane defined by p0 and r0, so a more accurate representation (generated by the SX1301 script) would show two lines representing the plane intersecting the front and back of the data-cube.
The line slope in each of two 2D projections plotted against Avg Delta Mass – peak Count and log (Rank+1) - may be used to estimate p0 and r0, respectively. Resolution of 100 seems to be more than sufficient for both parameters.
A detailed study of Figure 5 shows that a few decoy peptides achieve surprisingly high peak Count with low delta-masses, such that it seems hard to believe they can be incorrect IDs. One possible explanation is that these are actually correct peptide IDs that happen to be missing from the target sequences, but appear as a decoy by random chance. But our unpublished analysis suggests this may not be the case.
Instead, we found that many such decoys are not truly “correct” peptide IDs per se, but rather they are sequences with high sequence homology to the presumed correct ones. Notably search engine similarity scores reflect sequence homology, not identity, so they survive the first-level filtering of being among the top-N candidate peptides. Long peptides also match more raw fragments than short ones even if subsequences don’t match. This is a subtle but important distinction for deep analysis.
Implicit decoys constructed with peptide terminus modifications (e.g. ETD mods), which shift b- and y-ion series independently, frequently show up as lower ranked versions of correct peptides, with comparable S-scores, similar peak Counts, but >>1amu precursor delta-mass.
A related issue is the presumed hidden -incorrect IDs presumably mirrored by the explicit decoys. We hypothesize that, per standard target-decoy theory, there is an approximately same number of similar-but-not-quite-correct target peptide IDs with comparable degree of sequence homology.
Figure 7 shows a clearly large population of likely correct IDs with raw rank=1 [i.e. log (Rank+1)=0.693] and low Avg Delta Mass. There is a generally linear boundary shown with representative line of slope -r0.
The plot shows that using a non-linear boundary or using different boundaries for rank=1 vs. rank>1 will yield higher peptide IDs for a given overall FDR. That may or may not make sense depending on the objective of the data analysis. Adding more complexity increases possibility of over-fitting.
Distribution of S-score and FDR
Figure 8 shows the superimposed S-score distributions of decoys (yellow) and non-decoys (gray). Left of the dashed line, the decoy and non-decoy distributions are evenly matched (the yellow mostly match the gray behind it), suggesting mostly random incorrect IDs. Right of the solid line, non-decoys are likely-correct IDs. In-between is the transition region that may require additional discriminators to resolve.
The SX1301 MUSE script is designed to run automatically within the SORCERER system using its GEMYNI software platform. The search requires a standard 1:1 target-decoy distribution (i.e. for “explicit” decoys) and the top 200 or more XCorr peptides. Clearly, Sorcerer Score requires non-corrupt data and valid search results on which it can improve. Search results with significant data quality problems, which can be up to 10% of proteomics experiments, cannot be fixed by the SX1301 script or any other computational approach.
For integration into Trans-Proteomic Pipeline and compatible workflows, the S-score is used to derive an equivalent Peptide Prophet discriminant score. The current script generates a new search output file (SQT) whose top peptides are the top S-score peptides with their original XCorr, and whose second peptide is a dummy entry designed to produce the desired discriminant score once the standard Peptide Prophet calculations are performed.
Sorcerer Score can also be used as a standalone capability for targeted analysis, such as for clinical biomarker discovery and deep pathway analysis at the peptide level. In a nutshell, intermediate data files generated by SX1301 may be extracted and re-analyzed off-line, including quantitation and PTM analysis. Unlike closed “programs”, the flexible GEMYNI scripting platform enables capable developers to perform specialized deep analytics. Internal data files are stored in text format suitable for scripting and data-mining.
The foundation of target-decoy is captured in this math puzzle: Suppose someone tosses 120 coins up in the air and 20 come up tails. How would you explain this?
If you assume there are two kinds of coins - two-headed coins (correct IDs with 100% heads) and fair coins (random incorrect IDs with 50%:50% heads: tails) - then there would be ~80 two-headed coins and ~40 fair coins, the latter suggested by 20 tails.
More generally, ʻNʼ coins with ʻDʼ tails means roughly ʻ2*Dʼ fair coins and ʻN-Dʼ two-headed coins. Therefore, after all decoys are removed, the estimated FDR is “D/ (N-D)” in the remaining population. For our example, we remove the 20 tails to leave 100 coins, and expect to have 20 fair coins hidden among 80 two-headed coins (i.e. FDR~20%).
In proteomics, if 120K candidate peptide IDs, perhaps after some rigorous filtering, contains 20K decoys, then the 100K non-decoys are expected to have an estimated FDR ~20%.
The key point is the implied one-to-one correspondence between decoys you see and the incorrect non-decoys you donʼt. In other words, decoys are merely proxies, like shadows that represent objects hidden behind a wall. Any rigorous post-search filter must remove equal (or greater) numbers of non-decoys vs. decoys.
In contrast, a non-rigorous filter can cheat by preferentially removing decoys to make FDR look better. In the above example, if someone steals 19 tails to leave 101 coins with 1 tail, then FDR suddenly looks better at 1% even though the true FDR remains 20%. Opaque or complex algorithms make it easy to hide such problems.
This point illustrates the difference between non-rigorous “demo” analytics and rigorous analytics. They generally agree on low-noise data but disagree on high-noise or corrupt data, where demo analytics is designed to report great results no matter what. Researchers, who don’t understand the distinction may benchmark two tools with a simple experiment, conclude they have similar quality, and proceed to do real experiments with the lower-priced demo software, putting their research at risk.
Sorcerer Scoreʼs rigorous post-search filtering can be visually checked by noting the nearly 1:1 correspondence for low S-score distributions in Figure 8.
One subtle point that can affect deep analysis, depending on the dataset, is that the distribution of target sequences to decoys for incorrect IDs is not exactly 50%:50%. In our unpublished analysis, we have observed variations from 49.5%:50.5% to 50.5%: 49.5% that arise from different types of decoys (reversed vs. scrambled vs. peptide-terminus modifications) and the depth of top scores kept (top-1 vs. top-500). Although the variation sounds minor at first, the effect can be >10% in the estimated number of correct IDs because the percentage of correct IDs can be a tiny portion of the overall population.
We strongly recommend pseudo-reversed sequences for explicit decoys,2 the default on SORCERER systems, for deep proteomics because of better matching target-vs-decoy precursor mass distributions.
Many researchers are unaware that reversing the sequence produces a different mass distribution because of the asymmetry of enzymatic digest.
A target proteinʼs amino acid sequence of “...KabcdeR...” yields the tryptic peptide “abcdeR”. A standard reversed sequence yields “edcbaK” with a different mass, but pseudo-reversed keeps the n-terminus intact to yield “edcbaR” with the same mass. This is especially important for targeted searches against a small protein sequence database, such as for pathway analysis, to be as close as possible to 50%-50% target-decoy distributions at the individual spectrum level.
Robust characterization of low-abundance and/or modified peptides and proteins is a revolutionary game-changer for molecular biology research. Sorcerer Score makes it possible for the first time with a simple yet scientifically rigorous methodology. Deep proteomics is fundamentally an analytics challenge that requires attention to subtle details.
None.
The author declares no conflict of interest.

©2016 Chiang. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.