MOJ
eISSN: 2374-6920


Research Article Volume 2 Issue 1
Department of Biological Sciences, Charles E. Schmidt College of Science, Florida Atlantic University, USA
Correspondence: Ramaswamy Narayanan, Department of Biological Sciences, Charles E. Schmidt College of Science, Florida Atlantic University, 777 Glades Road, Boca Raton, FL 33431, USA, Tel 15612972247, Fax 15612973859
Received: December 24, 2014 | Published: December 18, 2014
Citation: Narayanan R. Neurodegenerative diseases: phenome to genome analysis. MOJ Proteomics Bioinform. 2015;2(1):1-8. DOI: 10.15406/mojpb.2014.02.00033
Genome-Wide Association Studies (GWAS) tools are an attractive starting point for disease target discovery. Genetic association evidence provides a strong link to disease phenotypes across diverse human DNAs. The National Center for Biotechnology Information (NCBI) Phenome-Genome Integrator tool (PheGenI) was used to identify genes associated with neurodegenerative disease phenotypes (NeuroGenes). Four hundred ninty-nine known proteins and 30 uncharacterized proteins were found to be associated with diverse neurological diseases including Alzeheimer’s, amyotrophic lateral sclerosis (ALS), epilepsy, multiple scelerosis, stroke and Parkinson’s. The NeuroGenes also were associated with other diseases and associated disorders. Using diverse bioinformatics and proteomics tools, an atlas of the NeuroGenes was created to facilitate rational biomarker and drug target discovery. NeuroGenes protein expression was detected in diverse body fluids. The NeuroGenes were categorized into functional classes of proteins using protein motif and domains analysis tools. Expression Quantitative Trait Loci (eQTL) analysis identified single nucleotide polymorphisms (SNPs) associated with mRNA expression across diverse sets of samples from Alzheimer’s disease, Parkinson’s disease, brain infarction, multiple sclerosis, schizophrenia, stroke, mental retardation and neurological cancers. Thirty uncharacterized proteins encompassing structural protein, translation initiation factor, GDP/GTP exchange factor (GEF), transmembrane proteins, tubulin/histone-binding and a lysosomal hydrolase were identified as putative proteins for dementia, Parkinson’s, neurobehavior, multiple scelerosis and ALS. These results may provide new biomarkers for neurological diseases.
Keywords: Neurological Diseases; Bioinformatics; Proteomics; Genetic Association; Body Fluids; Phenome Analysis; Human Genome; Human Proteome Map; Druggable Targets; Biomarkers; NeuroGenes
ClinVar, clinical variations; FDA, federal drug administration; eQTL, expression quantitative trait loci; GAD, genetic association database; GtEx, genotype-tissue expression; HapMap, haploid map; HGNC, human genome nomenclature committee; MOPED, model organism protein expression database; PheGenI, phenotype-genotype integrator; ORF, open reading frame; RefSeq, reference sequence; SNP, single nucleotide polymorphism
Neurodegenerative diseases comprise complex sets of disorders which strike primarily in mid- to late-life; their incidence is expected to soar as the population ages. According to the Alzheimer’s Association’s statistics, five million Americans suffer from Alzheimer’s disease. The risk of developing Alzheimer’s disease increases with age, and about one in 8 people over the age of 65 is affected by Alzheimer’s disease. It is the most common form of mental decline or dementia in older adults.
Further, 1 million people suffer from Parkinson’s disease, 400,000 from multiple sclerosis (MS), 30,000 from amyotrophic lateral sclerosis (ALS or Lou Gehrig’s disease), and 30,000 from Huntington’s disease.1 Novel molecular targets are needed to develop a better understanding of these diseases for diagnosis and treatment.
Genome-Wide Association Studies (GWAS) offer clues to assessing risk of development, susceptibility and cures to diverse neurodegenerative diseases.1–10 Identifying molecular targets from the GWAS databases offers an improved starting point since a great deal of patient-derived association evidence is readily available. Further, information related to the Copy Number Variations (CNV), gene expression at mRNA levels and the Expression Quantitative Trait Loci (eQTL) across the 1000 Genomes Project from these databases establishesa stronger correlation of a gene target in the pathology of the disease.
The National Center for Biotechnology Information (NCBI) Phenome-Genome Integrator (PheGenI) tool merges National Human Genome Research Institute (NHGRI) GWAS data with several databases, including Gene, database of Genotypes and Phenotypes (dbGaP), Online Mendelian Inheritance in Man (OMIM), the Genotype-Tissue Expression project (GTEx) and the Single Nucleotide Polymorphism database (dbSNP). This phenotype-oriented resource facilitates follow-up studies from GWAS and allows prioritization of variants.11 Increasingly, bioinformatics and proteomics approaches are becoming important to target discovery for complex diseases such as neurodegenerative diseases.12–16
The availability of protein expression datasets from the Multi Omics Protein Expression Database (MOPED),17 the Human Proteome Map18 and the Proteomics DB19 for the human proteome has greatly facilitated the target validation process for biomarker discovery. These datasets encompass protein expression data from diverse tissues as well as body fluids.
Using a streamlined bioinformatics and proteomics approach, we have recently demonstrated mining of the human genome for target discovery in diseases including cancer and diabetes.20–22 In this study, the PheGenI tool was used to develop an initial list of genetically associated single nucleotide polymorphic (SNP) loci with neurological traits. The associated genes (NeuroGenes) were subjected to comprehensive bioinformatics and proteomics analyses (n=499). The NeuroGenes were organized into functional classes of proteins such as enzymes, transporters and extracellular proteins and expression in diverse body fluids was established using protein expression databases. Functional annotation of thirty uncharacterized proteins predicts putative targets for further studies to aid in neurodegenerative disease research.
Genome analysis
The genome analysis was performed using the Genetic Association Disease (GAD) database,23,24 the UCSC Genome Browser from the University of California Santa cruz,25 the Ensembl Genome Browser,26 the National Center for Biotechnology Information, NCBI Gene Expression Quantitative Loci Browser, eQTL11 and the Genotype-Tissue Expression Project, GTEx.27 The GAD database was downloaded and stored locally. Excel advanced filtering option was used to functionally annotate neurologically associated genes and a separate working database was generated for further manipulation.
Transcriptome analysis
The transcriptome analysis of the uncharacterized proteins was undertaken using the NCBI-UniGene, SAGE Digital Anatomical Viewer28 and the Array Express.29
Proteome analysis
The proteome of the uncharacterized ORFs was analyzed using the UniProt Knowledge Base, UniProtKB,30 the Swiss Institute of Bioinformatics (SIB) ExPASy Server’s Protein Database (PDB) and post-translational modification sites at ExPASy.31 Protein motifs and domains were analyzed using the NCBI Conserved Domain Database, CDD,32 the PFAM,33 ProDom,34 InterProscan4,35 HMMER36 and Signal P Server.37 Protein expression batch analysis was performed using the Human Protein Atlas, HPA,38 the Multi Omics Proteins Expression Database, MOPED,39 the Human Protopedia Reference Database, HPRD40 the Human Proteome Map18,41 and Proteomes database, proteomics dB.19 For the batch analysis, a comma-separated values (CSV) file of the NeuroGenes list was first generated for use with the protein expression analyses tools indicated above.
Knowledge-based data mining
Protein and genome knowledge databases used included GeneCards,42 the MalaCards43 Online Mendelian Inheritance in Man (OMIM), Human Genome Nomenclature Committee, HGNC,44 Gene Ontology, Amigo,45 NCBI SNP database, the NCBI Phenome-Genome integrator, PheGenI,11 the Expression Quantitative Trait Browser, eQTL,27 the NCBI Clinical Variations database, ClinVar,46 the KEGG pathway and the DAVID functional annotation tool.47 For PheGenI tool the P-value was set at values <10-5 and the R-Squared at 0.3. For Clinical Variations, the variation filter was set at pathogenic or risk factor associated single nucleotide polymorphisms.
GeneALaCart batch analysis
The GeneALaCart Meta analysis tool from the GeneCards (LifeMap Discovery) was used to batch analyse the NeuroGenes for gene names, aliases and descriptions, gene ontology, protein motif and domains, pathways and interactome analysis and drug bank hits.
Data analysis
The entire database of GAD, Human Protein Atlas, HPA and UniGene was downloaded and the Excel filtering tool was used to scan for the ORFs. Batch analysis of the ORF database was performed for canSar, the MOPED, the DAVID annotation tool, the Human Proteome Map, Proteomics dB, the PheGenI and the eQTL browser.
All of the bioinformatics mining was verified by two independent experiments. For the bioinformatics tools used in the study, the basic parameters indicated by the tool’s browser were used. Only statistically significant results per each tool’s requirement are reported. Big data from the experiments was first generated as tab-separated values file and was imported into a working Excel document. The Excel data was filtered using the advanced filtering options to create annotated clusters. Prior to using a bioinformatics tool, a series of control query sequences was tested to evaluate the predicted outcome of the results.20–22,48
Neurodegenerative disease-associated traits
Neurologically relevant gene-related information exists in different databases, which complicates efforts to identify targets for further studies.49–51 In order to facilitate mining the human genome for neuro targets, the NCBI PheGenI was analyzed for traits associated with nervous system diseases.
Polymorphic traits, SNPs (n=1,034) associated with diverse neurological diseases and disorders were identified (Figure 1). These SNPs were present in 499 genes encompassing both the known and novel, uncharacterized protein open reading frames (ORFs). Alzheimer’s Disease, mental competency, multiple sclerosis and Parkinson’s disease harbored the largest number of polymorphic SNPs and associated genes (Supplemental Table S1).
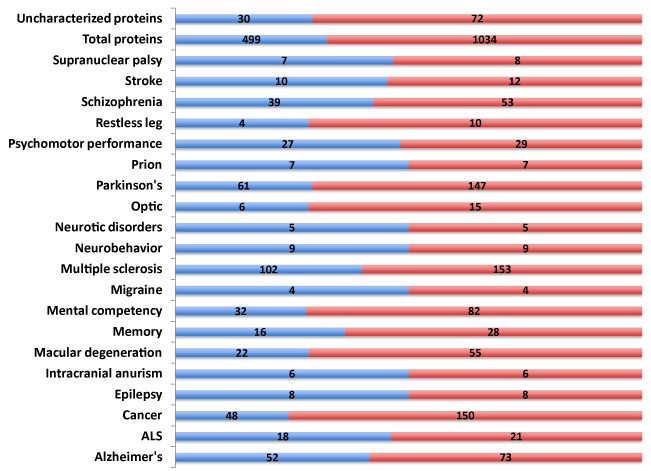
Reasoning that a comprehensive knowledgebase of the 499 genes associated with neurological disease (NeuroGenes) would facilitate target discovery, a comprehensive bioinformatics and proteomics analysis was undertaken. The GeneCards and DAVID Functional Annotation Meta Analysis tools were used to develop the knowledgebase, which encompasses protein identifiers, gene ontology, summary of function, protein motif and domain related information and pathways (Supplemental Table S2).
Neurogenes associated with diverse diseases
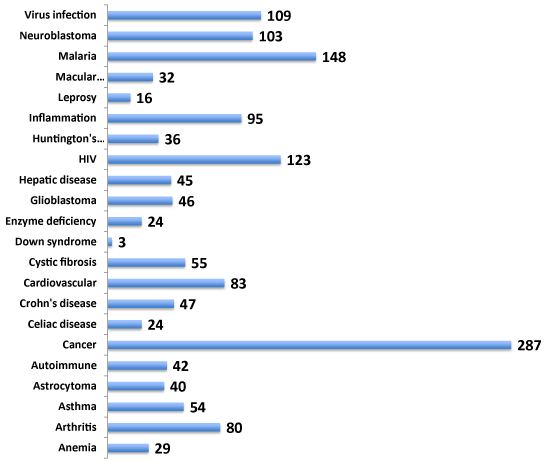
Gene function is often involved across multiple cell types eliciting cell type-specific networks of gene interactions. Identification of gene targets related to multiple diseases offers an advantage of benefitting the drug discovery rationale for therapy for numerous diseases and related disorders. Hence, the involvement of NeuroGenes both in neurologically related and unrelated diseases was next investigated from the NCBI PheGenI data (Figure 2). The NeuroGenes SNPs were also found to be associated with diverse diseases including allergy, cancer, cardiovascular, inflammation, macular degeneration and infections (Supplemental Table S3). Cancer (including astracytoma, glioblastoma and neuroblastoma) had the largest number of gene-associated evidence (n=189). These results underscore the complexity of genes’ involvement in multiple diseases.
Protein expression of neurogenes in body fluids: baseline analysis
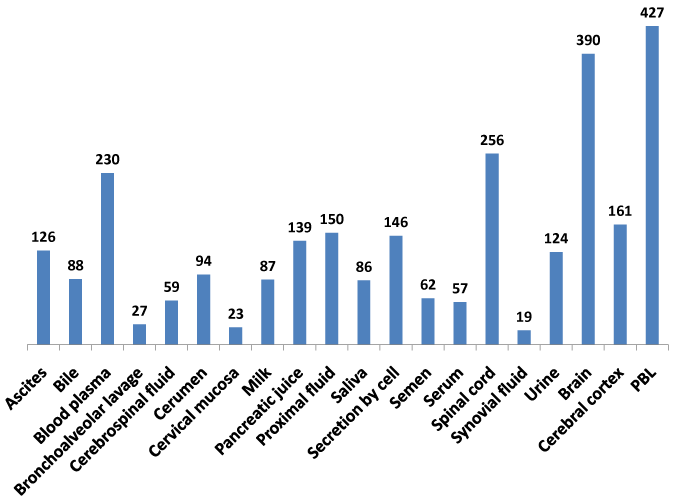
The recent availability of the human proteome map dataset for protein expression in multiple tissues and body fluids18,19,39 enabled this study to develop a comprehensive baseline protein expression dataset on the NeuroGenes (Figure 3). A composite protein expression dataset was generated using the MOPED, the Human Proteome Map and the ProteomicsDB databases. The NeuroGenes protein expression was detected in diverse body fluids including ascites, cerebrospinal fluid, milk, saliva, serum, semen and urine. Most of the Neurogenes were detected in the brain, spinal cord tissues and peripheral blood lymphocytes (Supplemental Table S4). This baseline analysis for protein expression in normal human body fluids establishes a framework for biomarker screening efforts for diverse neurological diseases.
Gene ontology of neurogenes: cell location, function and process
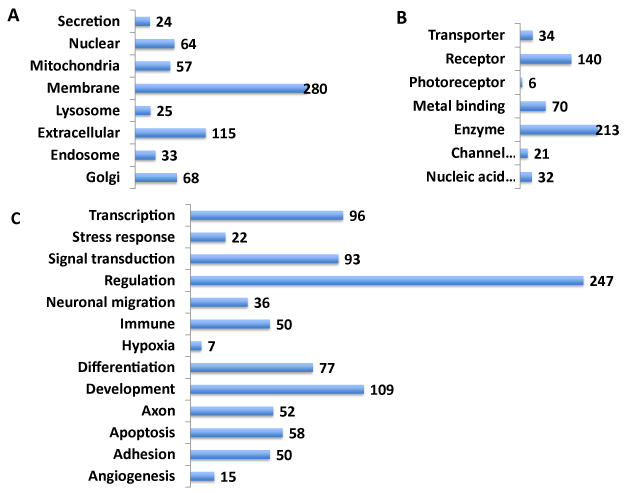
To add to the NeuroGenes knowledgebase, the Gene Ontology was investigated next using the GeneALaCart Meta Analysis tool (Figure 4). The proteins were classified into cellular location (Panel A), function (Panel B) and processes involved (Panel C). The largest number of proteins was found to be membrane bound (280/499). In addition, extracellular, secreted, nuclear/mitochondrial/Golgi as well as cytoskeletal proteins were inferred (Panel A; also Supplemental Table S5). The NeuroGenes encompassed druggable targets including channel proteins, transporters, receptors and enzymes (Panel B). Enzymes constituted the largest druggable class of proteins (213/499). The NeuroGenes were implicated in diverse biological processes such as angiogenesis, apoptosis, axon, differentiation and development, immune function, neuronal migration and signal transduction (Panel C). The largest number of NeuroGenes was associated with regulation of gene expression (247/499).
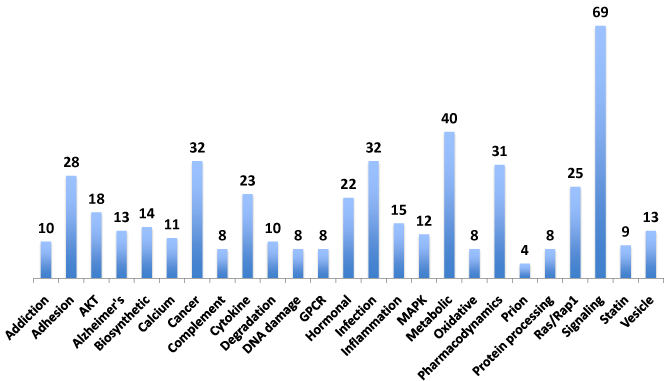
Neurogene pathways
Elucidating the pathways implicated in the expression of the NeuroGenes is essential to deriving functional knowledge for further studies. The pathway-related information was inferred from the KEGG and BioCarta pathway analyses tools (Figure 5). Diverse pathways such as addiction, AKT/MAPK signaling, Alzheimer’s, inflammation, infections, metabolic, prion, STAT signaling and vesicular trafficking were involved with the NeuroGenes (Supplemental Table S6).
Neurogene protein class and families
Hints regarding function were next investigated using protein motif and domain analyses tools, the HGNC motifs and domains, UniProt Domains, the InterPro, the Signal P server, the Protein Family, PFAM and the DAVID functional annotation tool (Figure 6). The 289 NeuroGenes were classified into adhesion molecules, ligand and metal binding proteins, enzymes, receptors, transporters, channel proteins, immunoglobulins and transcription factors (Supplemental Table S7). These results further verified the possible druggable nature of many NeuroGenes.

Expression quantitative trait loci analysis of the neurogenes
An expression quantitative trait locus (eQTL) represents a marker (locus) in the genome in which variation between individuals is associated with a quantitative gene expression trait, often measured as mRNA abundance. The three components critical to validating eQTL results include i) a SNP marker; ii) the gene expression levels, as measured by a probe or sequence information; and iii) a measure of the statistical association between the two in a study population, such as the P-value. The eQTL browser provides an approach to query the eQTL database.52
The eQTLs can be cis, where the genotyped marker is near the expressed gene, or trans, in which the genotyped marker is distant from the expressed gene either in the same or on another chromosome. Currently only the cis-eQTLs53 are available. In order to establish neurological disease-associated eQTL results for the NeuroGenes, the PheGenI tool was used to batch analyze the SNPs associated with the NeuroGenes. Genotypes for the NeuroGenes were selected for exons, introns, near gene and Untranslated Region (UTR).
From these population studies, 18 NeuroGenes were found to have eQTL association with diverse neurological diseases (Table 1 Included as supplementary). The SNPs for these 18 genes were present in diverse human DNAs from the 1000 Genomes Project. The diseases showing associated eQTL genes included Alzheimer’s disease (HLA-A, MAPK8IP1, MAPT), dementia (ITIH4), multiple scelerosis (CLECL1, FAM119B, HLA-DQB1, HLA-G and RPS6KB1), natriuretic peptide (NFIA), neuroblastoma (BARD1, RPS6KB2), neural tube defect (MTHFR), Parkinson’s disease (LRRC37A, MAPK8IP1 and MAPT), schizophrenia (AHI1, HLA-DQA1 and HLA-A), stroke (HLA-DQA1), prion disease (MAPT and HLA-DQB1) and retinoblastoma (HLA-DRA).
The ribosomal protein S6 kinase, 70kDa, polypeptide 1 (RPS6KB1) was associated with diverse neuronal cancers including malignant peripheral nerve sheath tumor, neuroendocrine tumor, glioblastoma multiforme, malignant glioma, astrocytoma, neuroblastoma and neuronitis. The Genotype-Tissue Expression project (GTEx) portal analysis showed that 13/14 eQTL genes were expressed in all tissues analyzed (Supplemental Table S8).
The NCBI ClinVar dataset showed clinically relevant SNPs for frontotemporal dementia, systemic lupus, diabetes Type 1 and Type 2, celiac disease, developmental delay, Grave’s disease, hereditary neuroblastoma, risk factors for psoriasis susceptibility 1, and susceptibility to HIV-1 viremia and Schizophrenia 9 (Table 1 Included as supplementary).
Uncharacterized neurogenes: novel targets
Recently we have mined the human proteome for uncharacterized proteins to enable novel disease target discovery. Targets relevant to cancer and diabetes were identified.21,22,48,54 Reasoning that the 30 uncharacterized proteins, that showed strong genetic association with neurological diseases may offer novel biomarker and target potential, a comprehensive analysis was undertaken (Table 2 Included as supplementary). A structural protein, C1orf125 containing axonal dynin light chain was found to be associated with multiple sclerosis, Huntington’s disease and diabetes Type 2.55,56
Six Parkinson’s disease-associated novel proteins were also identified. These included FAM47E (lncRNA transcription co factor), KIAA0427 (translation initiation factor), KIAA1267 (histone binding), TMEM175 (lysosomal transmembrane protein), C9orf72 (GDP/GTP exchange factor) and a novel enzyme, TMEM55A (transmembrane hydrolase).
The C20orf196 (uncharacterized), FAM110C (lncRNA alpha-tubulin binding protein) and C9orf5 (transmembrane glycoprotein) were associated with neurobehavioral manifestations.57,58 Two of the ORFs were associated with carotid artery diseases, C9orf93, a myosin-related nucleotide binding protein55 and C20orf196 (uncharacterized protein).58
A GDP/GTP exchange factor (GEF) showed restricted expression in the adult brain and peripheral blood lymphocytes and was associated with amyotrophic lateral sclerosis, Huntington’s, frontotemporal dementia and Parkinson’s.59–62 A repeat expansion in the C9orf72/GEF gene has recently been identified as a major cause of familial and sporadic frontotemporal lobar degeneration, Huntington’s, Parkinson’s and amyotrophic lateral sclerosis. A membrane testis-specific basic protein, C6orf10, showed association with multiple sclerosis,63 asthma64 and rheumatoid arthritis65 (Supplemental Table S9). Additional bioinformatics details on these uncharacterized NeuroORFs are shown in the (Supplemental Table S10). These novel proteins offer opportunities for further development of novel diagnostics and therapeutics.
Therapeutic value of the neurogenes
Drug hits for genes (including chemical compounds and Federal Drug Administration (FDA) approved drugs) are available in diverse drug-related databases. Reasoning that mining for drugs implicated with the pathways associated with the NeuroGenes might offer a framework for understanding mechanism, the NeuroGenes were batch analyzed using the GeneALaCart Meta Analysis tool for drug hits in the drug banks.
Numerous FDA-approved drug hits and well as other compounds which target the NeuroGene’s pathways were identified including statins, Cyclosporine A, Donepezil, Rosiglitazone (Alzheimer’s disease, cognition), antivirals such as ritonavir and Ganciclovir, hormones including Estrogen and Testosterone, Verapamil, Isradipine (gating inhibitor), Diltiazem (Channel blocker), Gemtuzumabozogamicin, Cisplatin and Methotrexate (antineoplastics) and antibiotics such as rapamycin, Tunicamycin (Supplemental Table S11) for complete details).
Diagnosis and therapy of complex neurodegenerative diseases requires novel molecular targets. Current drug targets for Alzheimer’s disease, Parkinson’s, Huntington’s disease and ALS revolve around functional classes of proteins such as protein aggregation, mitochondrion proteins, N-methyl-d-aspartic acid (NMDA) receptors, voltage gated calcium channels (VGCCs), neuronal nitric oxide synthase (nNOS), oxidative stress from reactive oxygen species, and DNA damage repair enzymes.66–68
Individuals with neurodegenerative diseases such as Parkinson’s or Alzheimer’s are currently benefiting from drugs that target a single gene. However, the single agent treatment modality is inefficient. Increasingly it is becoming a paradigm in drug discovery to focus on drug candidates that affect multiple neural and biochemical targets for the treatment of diverse associated disorders such as cognition impairment, motor dysfunction, depression and neurodegeneration.66,69–71
Identification of molecular targets from unbiased GWAS databases is likely to contribute to better therapeutic target discovery for neurodegenerative diseases.2,72 Further, such targets are likely to be more relevant to diverse populations because of strong genetic association evidence across the global population DNAs. Utilizing a similar rationale, pancreatic cancer targets were recently identified.20
In this study, neurologically associated traits were enriched based on GWAS evidence and a functional annotation of the classes of proteins was established. The 499 NeuroGenes thus identified showed overlapping association evidence with other diseases and disorders including allergy, cancer, cardiovascular, infections, inflammation and autoimmune disorders. Interestingly, a significant number of these proteins were detected in diverse body fluids such as blood, cerebrospinal fluid, milk, urine, saliva or semen. A strong correlation of protein expression data was seen when using different protein expression databases such as the MOPED and Proteomics DB. This is encouraging as far as the specificity of prediction is concerned, since the tissue samples for these studies came from different patients. These results also provide baseline expression data for these proteins to further the development of novel biomarkers for diverse neurological diseases.
Using gene ontology and protein motif and domain analysis, the NeuroGenes were further categorized into functional classes of proteins. The database of NeuroGenes was sorted into protein classes encompassing druggable genes including enzymes, G protein coupled receptors, transporters, transcription factors and channel proteins.73 This should aid in the prioritization of efforts to validate target genes.
The eQTL analysis of the NeuroGenes led to the identification of 18 genes showing strong association with ALS, Alzheimer’s, cancer, multiple sclerosis, Parkinson’s and schizophrenia. Of interest was the discovery among those identified of RPS6KB1, a member of the ribosomal S6 kinase family of serine/threonine kinases. The encoded protein responds to mTOR (mammalian target of rapamycin) signaling to promote protein synthesis, cell growth, and cell proliferation. This gene is a risk factor for a great number of tumor types including malignant peripheral nerve sheath tumor, neuroendocrine tumor, glioblastoma multiforme, malignant glioma, astrocytoma and neuroblastoma, lymphoma, breast tumors, non-small cell lung carcinoma, in addition to Alzheimer’s disease and multiple sclerosis.74–79 Consistent with these results, the Catalogue of Somatic Mutation in Cancer database, COSMIC80 was found to harbor a large number of somatic mutations in the protein kinase domain (substitution missense). Moreover, mouse insertional mutagenesis experiments support RPS6KB1 as a driver cancer-causing gene.81,82 This kinase offers a novel biomarker and drug therapy opportunity for diverse cancers.
This study also identified 30 uncharacterized proteins with association evidence in multiple sclerosis, Parkinson’s disease, Huntington’s disease, amyotrophic lateral sclerosis and frontotemporal dementia. The protein classes encompassed included transmembrane glycoprotein, GDP/GTP exchange factor, transcription factor, myosin/tubulin binding proteins, LDL and nuclear receptors, transporters and a lysosomal transmembrane enzyme. Six of the 30 proteins were associated with Parkinson’s disease. It is tempting to suggest that these may offer a fingerprint for Parkinson’s disease. Additional verification is warranted.
Analysis of the drug banks identified FDA-approved drugs and as well as compounds implicating diverse biochemical pathways involved with the NeuroGenes. These drugs and compounds provides a starting point for use in model systems and for improved Structure Activity Relationship (SAR) studies in the future.
Identification of proteins as genetically linked using the GWAS dataset provides a strong rationale for lead prioritization in biomarker and therapeutic target research. Such an approach is likely to lead to the development of more relevant targets for further studies.
The results presented in this study demonstrate that mining the phenome-genome association databases can be an effective approach for disease biomarker and putative target discovery for further study. The 30 novel proteins characterized in this study shed more light on the Dark Matter proteome and offer new opportunities for understanding diverse neurological diseases. The atlas of the NeuroGenes generated in this study can provide a starting point for target verification for biomarker potential and eventual drug therapy use.
This work was supported in part by the Genomics of Cancer Fund, Florida Atlantic University Foundation. I thank Dr. Stein of the GeneCards team for generous permission to use the powerful GeneALaCart tool; Dr. Montague, Kolker Laboratory of the MOPED Team for batch analysis of the ORFs and the Human Proteome Map and ProteomesDB for the datasets. I thank Jeanine Narayanan for editorial assistance.
The author declares no conflict of interest.

©2014 Narayanan. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.