MOJ
eISSN: 2374-6920


Research Article Volume 1 Issue 5
Department of Biological Sciences, Florida Atlantic University, USA
Correspondence: Ramaswamy Narayanan, Department of Biological Sciences, Charles E. Schmidt College of Science, Florida Atlantic University, 777 Glades Road, Boca Raton, FL 33431, USA, Tel 15612972247, Fax 15612973859
Received: December 05, 2014 | Published: December 10, 2014
Citation: Narayanan R. Ebola-associated genes in the human genome: implications for novel targets. MOJ Proteomics Bioinform. 2014;1(5):139-144. DOI: 10.15406/mojpb.2014.01.00032
Ebola Virus Disease (EVD) is a major healthcare challenge facing the globe today and if left unchecked could become a pandemic. A limited knowledgebase exists about the Ebola virus with no U.S. Federal Drug Agency (FDA) approved drugs. Ebola-specific proteins, antibodies and vaccines are being explored currently for therapy. In an attempt to first develop an understanding of the human proteins involved in the EVD and potentially create a pipeline of targets for evaluation, the human genome was mined for EVD association using 1) genetic association, 2) disease-oriented knowledge database text mining, 3) transcriptome-based Meta Analysis and 4) pathway enrichment analysis. Forty-five human proteins (29 known proteins and 16 novel previously uncharacterized proteins) were identified which were associated with EVD. Mining the proteomic expression databases revealed the detection of these 45 proteins in diverse body fluids. Detailed bioinformatics and proteomic analyses of these EVD associated proteins shed light on pathways, nature and class of the proteins and their association with diverse diseases including other infectious diseases. Mining the drug banks for association with the 45 genes reveals putative drugs including anti-neoplastics, anti-inflammatory drugs, leukotrienes, interferons, anticoagulants, nucleoside analogues, retinoic acid and statins to add to the currently meager options for therapy and supportive care. Putative targets of interest include a chemokine (C-C motif) ligand 8 (CCL8), enzymes (indoleamine 2,3-dioxygenase 1|IDO1, cytochrome c oxidase| COA6) and Short-chain dehydrogenase/reductase, SDR) and members of the receptor family, killer cell immunoglobulin-like receptor, cytoplasmic tail (KIR). Using knockout technology, the 45 proteins identified in this study can be rapidly verified for therapeutic potential. The association of these proteins with other diseases potentially expands the scope to diverse use. The results presented in this study provide a further example of harnessing the human genome to identify disease relevant proteins for subsequent research and development.
Keywords: genome to phenome, genetic association, hemorrhagic fever, virus, human genome, pathogen, host targets, proteome analysis, motif and domains, pharmcogenomics
AIDS, acquired immune deficiency syndrome; ClinVar, clinical variations; EVD, Ebola virus disease; eQTL, expression quantitative trait loci; KIR, killer cell immunoglobulin receptor; NCBI, national center for biotechnology information; PheGenI, phenome genome integrator
Both Ebola and Marburg viruses belong to the Filoviridae family of viruses and cause highly lethal hemorrhagic fevers. Ebola virus contains a single-stranded, non-infectious RNA genome.1 The Ebola virus genome comprises seven genes including 3’-UTR-NP-VP35-VP40-GP-VP30-VP24-L-5’-UTR.2,3 There are at least five different species of Ebola virus and the infection has a mortality rate of over 90%, whereas the Marburg virus infection is associated with 20-90% mortality rate.1,2,4,5 Natural outbreaks of the Ebola virus have been reported in various parts of Africa.1,5–7 The current outbreak in West Africa, whose first cases were noted in March 2014, is the largest and most complex Ebola outbreak since the discovery of the virus in 1976 (World Health Organization, Fact sheet No. 103).
According to a U.S. Centers for Disease Control and Prevention Report (Nov. 2014), in this outbreak of Ebola Virus Disease (EVD), patients showed abrupt onset of fever and symptoms, typically 8-12days after exposure. Because of various non-specific symptoms, the early onset of EVD is often mistaken for other infectious diseases including malaria, typhoid fever, meningococcal infections and other bacterial infections.1
The pathogenesis of EVD primarily involves deregulation of the host’s immune response.2 Entry of the virus is via the mucous membranes, breaks in the skin or by ingestion. Numerous cell types are targets for infection including monocytes, macrophages, endothelial cells, dendritic cells, hepatocytes, epithelial cells and fibroblasts.8,9 EVD is associated with a massive release of pro-inflammatory cytokines, vascular leakage and impairment of clotting, resulting in multi-organ failure, shock and death.2,10,11
Currently, no FDA-approved vaccines or therapies are available and clinical management of the disease largely relies on supportive care of the associated complications such as fluid replacement, nutritional support, pain control, blood pressure maintenance and treatment of secondary bacterial infections (U.S. Centers for Disease Control and Prevention Report (Nov. 2014). Several investigational drugs including vaccines, antibodies, convalescent serum and siRNAs targeting Ebola virus proteins are currently being explored in clinical trials.12–14 However, new molecular targets and a rationale for the immediate use of drugs already approved for other indications are needed to overcome the current epidemic.
The completion of the human and pathogen genomes in the recent years has opened new avenues for diagnosis and therapy of infectious diseases.15,16 Whereas the pathogen genome and the proteins encoded by the etiological agents offer diagnostic markers and antibodies, vaccines and inhibitors (nucleic acids and small molecular compounds), increasingly the host genome is providing valuable hints to therapy.15–17 The Genome Wide Association Studies (GWAS) across the 1,000 and 10,000 genome projects is increasingly providing genome datasets for candidate genes identification, response to therapy prediction (pharmacogenomics), susceptibility or resistance to infection, and adsorption and entry of the pathogen into the host cell.15,17,18
The HIV field of research has provided a strong proof of concept for the importance of host cell genes in developing novel therapeutics. Resistance to HIV infections in homozygotes for C-C chemokine receptor type 5 (CCR5) has led to the development of drugs that would block this virus co-receptor.19–23 Similarly, based on other host cell protein polymorphism data, novel pharmacological inhibitors have emerged for diverse infectious diseases (e.g., cholera, malaria, tuberculosis, leprosy, hepatitis B), cancer and neurodegenerative diseases.17
In this report, by means of the disease and genetic association -oriented databases, the GAD, the MalaCards from the GeneCards and the Next Bio Transcriptome Meta analysis tool, 45 EVD-associated human genes (29 known genes and 16 uncharacterized ORFs) were identified. Bioinformatics and proteomics mining of these proteins led to data on functional class prediction, body fluids expression status, pathway mapping, association and clinical relevance with diverse diseases and drugs-related (including currently FDA approved for other indications) information. The results provide a starting point for host genome related target verification research.
The bioinformatics and proteomics tools used in the study have been described elsewhere.24–27 The following genome-wide association tools were used: the Genetic Association Database, GAD,28 the National Center for Biotechnology Information (NCBI) Phenotype-Genotype Integrator, PheGenI,29 the Expression Quantitative Trait (eQTL) GtEx browser,30 the Database of Genomic Variants, DGV,31 Clinical Variations and the ClinVar.32
Meta Analysis of the EVD-associated genes was performed using the Database for Annotation, Visualization and Integrated Discovery, DAVID v6.7 from the NCBI.33 The GeneALaCart (LifeMap discovery) from the GeneCards34 was used to batch analyze the query genes for gene names, protein IDs, motif and domain analysis, pathways and drug target discovery. The NextBio Transcriptome Meta analysis tool was used to identify most correlated tissues, drug interactions and gene perturbations.
The entire database of GAD and Human Protein Map, HPM35 was downloaded and the Excel filtering tool was used to scan for EVD-associated genes. Protein expression was established from the Batch analysis of the EVD-associated genes with the Multi Omics Protein Expression Database, MOPED,36 the DAVID annotation tool,33 the Human Proteome Map,37 Proteomics DB38 and the Human Proteins Reference Database, HPRD.39
The EVD association was verified using genetic association evidence from 1) the GAD and PheGenI, 2) disease-oriented knowledge bases (the MalaCards, Online Mendelian Inheritance in Man-OMIM), 3) transcriptome analysis from the microarray datasets (the NextBio Meta Analysis) and 4) pathway mapping (the GeneALaCart, DAVID).
All of the bioinformatics mining was verified by two independent experiments. Big data was downloaded two independent times and the output verified for consistency. Only statistically significant results per each tool’s requirement are reported. Prior to using a given bioinformatics tool, a series of control query sequences was tested to evaluate the predicted outcome of the results.
Identification of Ebola-associated proteins in the human genome
Reasoning that EVD-associated genes in the host genome might offer an understanding of mechanism and pathways involved for intervention, diverse disease oriented databases (the GAD, the PheGenI, MalaCards and the NextBio Meta Analysis transcriptome database) were scanned for association with EVD. Forty-four genes encompassing known genes (n=29) and uncharacterized ORFs (n=16) were identified which showed association with EVD (Table 1 Included as supplementary). The known genes included a chemokine C-C motif ligand 8 (CCL8), a complement component 1(C1QB), a transcription factor, ETS homologous factor (EHF), coagulation factor V (F5) an ion transporter, FXYD domain containing ion transport regulator 3 (FYDX3), a pancreatic glycoprotein, glycoprotein 2|zymogen granule membrane (GP2), an enzyme, indoleamine 2,3-dioxygenase 1 (IDO1), a cytokine regulatory factor, interferon regulatory factor 9 (IRF9) and diverse members of the immunoglobulin super family, the killer cell immunoglobulin-like receptors (KIR genes). In addition to their association with EVD, these proteins were also found to be associated with other bacterial and viral diseases such as CMV, encephalitis, hepatitis B, herpes simplex, HIV, influenza, leprosy, measles, swamp fever and West Nile virus. Moreover, associations with noninfectious diseases such as neurological, cardiac, cancer and metabolic disease were also seen with these known genes. The 16 ORFs identified in this study also were associated with EVD as well as other diseases and disorders (Figure 1).
The association of cytokines, immunoglobulin natural killer cells, EVD-associated infections and other unrelated diseases suggested involvement of overlapping mechanistic pathways (Supplemental Table S1). Hence, a detailed bioinformatics characterization of these 45 genes was undertaken.
Expression of EVD-associated human proteins in diverse body fluids
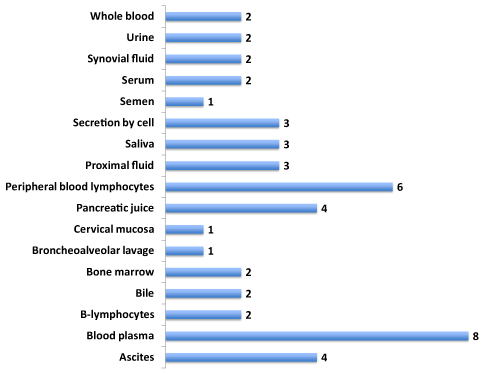
The protein expression datasets (MOPED, HPRD, Proteomics DB and the Human Proteome Map) from diverse human fetal and adult tissues as well as from body fluids for over 85% of the human proteins are available.36–38 These powerful databases have been used in numerous recent studies to establish expression of novel ORF proteins relevant to cancer, diabetes and neurodegenerative diseases.24,26,27 The eventual development of a diagnostic potential for the EVD-associated human proteins would necessitate ease of detection in the body fluids such as saliva, serum, urine etc. Hence the 45 EVD-associated proteins were batch analyzed against these expression databases. Figure 2 shows the expression data in diverse body fluids (Supplemental Data 2 for additional expression information). Expression of both the known proteins and the uncharacterized ORFs was seen in diverse body fluids including hematopoietic (blood, bone marrow, plasma, peripheral blood lymphocytes and serum), fluids (ascites, bile, pancreatic juice, proximal and synovial), saliva, semen and urine. Expression of these proteins was also seen in tissues relevant to EVD (MalaCards definition) including kidney, liver, skin and retina (Supplemental Data S2). In addition, expression was seen in B-cells, cytotoxic T-cells, monocytes, NK cells, spleen, platelets, and T-lymphocytes. The Human Proteome Map analysis of the EVD-associated proteins showed a highly selective expression profile for the individual members of the KIR gene family and the ORF proteins. Isoform-specific expression was seen with one member of the KIR gene family as monitored by the Proteomics DB. The killer cell immunoglobulin-like receptor, two domains, long cytoplasmic tail, 4, isoform 6 (KIR2DL4) showed the highest level of expression in the urine. These expression results underscore a high level of specificity of expression in distinct tissues and body fluids for the EVD-associated human proteins Supplemental Table S2).

Analysis of the dark matter of the human proteome associated with EVD
Sixteen uncharacterized ORFs were identified together with the known genes, which showed association with EVD as well as other diseases. In a series of recent reports our laboratory has demonstrated the diagnostic and therapy potential of these ORFs, termed the “Dark Matter” of the human proteome, in diverse therapeutic areas including cancer, diabetes and neurodegenerative diseases.25,26,40 Hypothesizing that these novel proteins may offer new molecular entities to further Ebola virus research, a comprehensive analysis of the ORFs was undertaken (Table 2 Included as supplementary). The ORF expression was detected in body fluids such as ascites, blood, bone marrow, peripheral blood lymphocytes and saliva. Protein motif and domain analysis of the ORFs revealed amino acid signatures for transcription factors, signal transduction proteins, enzymes and a secreted transmembrane signal peptide harboring protein (C10orf128). Two novel enzymes (C1orf31|cytochrome c oxidase assembly factor 6 homolog (S. cerevisiae) and an uncharacterized protein CXorf21|Short-chain dehydrogenase/reductase SDR) were among the EVD-associated ORFs. Expression of these two latter proteins was seen in bone marrow and blood plasma. Further, two novel regulatory proteins (C13orf34| aurora kinase A activator, BORA and C20orf117| suppressor of glucose, autophagy associated 1, SOGA1) were identified in the study (Supplemental Table S3 & S4). Two protein coding long intergenic RNAs (lincRNAs, C1orf42 and C1orf72) were among the 16 EVD-associated ORFs.
Gene ontology and pathways involved with the EVD-associated human proteins
In order to glean insight into the possible mechanistic functional and pathway information for the EVD-associated proteins, Gene Ontology and pathways-related information was inferred from the GeneALaCart (GeneCards) Meta Analysis tool and the DAVID functional annotation tool (Figure 2). The EVD-associated proteins encompass extracellular, membrane, mitochondrial and nuclear proteins. Functionally these proteins included antigens, cytokines, enzymes, Ig receptors, interferons, transcription factors and transporters. Diverse pathways relevant to infections including antigen processing and presentation, complement activation, inflammation, immune response, natural killer cell response, virus response and Akt/STAT/NF-kB/ERK signaling were implicated among the mechanisms for the EVD-associated proteins (Supplemental Table 5).
Drug hits in the databases for the EVD-associated human proteins
No effective treatment is currently available for EVD; fluid replacement therapy or simple antibiotic/anti-inflammatory drug therapy are being used with uncertain outcome.41,42 The mortality rate remains very high (85%). Reasoning that some of the EVD-associated human proteins might have hits in the drug banks, which might include currently FDA-approved drugs, the drug banks (HMDB, the Human Metabolome Database, Novoseek_Compounds, DrugBank_Compounds and PharmGKB, The Pharmacogenomics Knowledgebase) were screened for potential drug hits against the 46 EVD-associated proteins. The composite data shown in (Table 3 Included as supplementary) were obtained from the GeneALaCart Meta Analysis tool.
FDA-approved drugs as well as compounds involving the EVD-associated protein’s function including Cetuximab | Etanercept | Adalimumab | Abciximab | Gemtuzumab | ozogamicin | Trastuzumab | Rituximab | Tositumomab | Alefacept | Efalizumab | Natalizumab | Daclizumab | evacizumabSynagis | Avastin | leukotrieneb4 | heparin | Xigris | FIBRINOGEN | prostacyclin | sulfonamide | Tamoxifen | Simvastatin | Epinephrine | ceftriaxone | linezolid | tunicamycin | eltrombopag | hormonalcontraceptivesforsystemicuse | tamoxifen | Dexamethasone | infliximab | Chloramphenicol | interferonalfa-2a, | peginterferonalfa-2b | 5-Aza-2’deoxycytidine | retinoicacid) were associated with the EVD-associated known genes (Table 3 Included as supplementary). The 3-D protein structure information is available in the Protein Database (PDB) for nine of these known proteins; the PDB ID #s are shown in Table 3 (Included as supplementary). This should facilitate structure-based drug design approaches. For additional details on the implicated drugs see Supplemental Table S6. These results open up new opportunities for therapy of EVD as a basis of supportive care until effective vaccines are developed.
Landscape of EVD-associated human proteins across diverse diseases
Patients with EVD often show other opportunistic infections as well as other disorders. Hence, the disease-oriented databases were mined for association with other Ebola-associated diseases and disorders (Figure 3). The EVD-associated human proteins showed association evidence with immune disorders (autoimmune, AIDS), infections (viral, bacterial, parasitic) and hematopoietic disorders as well as neurological, inflammatory and eye diseases and cancer. The KIR family of genes showed association evidence across multiple diseases (Supplemental Table S7). The chemokine (C-C motif) ligand 8, CCL8 showed association with autoimmune diseases, viral infections, cancer and inflammation. Two genes (DEAD (Asp-Glu-Ala-Asp) box polypeptide 58, DDX58 and interferon-induced protein with tetratricopeptide repeats 2, IFIT2) were found to be uniquely associated with West Nile virus infections. A novel ORF, C2orf42 (a putative transcription factor|LincRNA) showed association evidence with malaria. These results underscore the complex involvement of EVD-associated proteins across a spectrum of diseases and disorders.

The KIR family of proteins in EVD
The KIR genes on chromosome 19 (19q34.1) encode the killer cell immunoglobulin-like receptors (KIR) and are expressed in natural killer cells and a subset of T-cells.43,44 To date 17 family members of the KIR gene including two pseudogenes have been identified with activation and inhibitory roles in NK cell function.45 The KIR genes are members of Ig-superfamily of type I membrane proteins and are highly polymorphic, binding to HLA class I alleles which drive NK cell function.46,47 Two members of the KIR activating family (KIR2DS1 and KIR2DS3) are shown to be associated with a fatal outcome in EVD.48 The genetic association evidence for the involvement of KIR family members in EVD and other diverse diseases and disorders is shown in Supplemental Table S8. Strong polymorphic association was seen for the distinct members of the KIR family in diverse bacterial and virus-related diseases. Clinically relevant pathogenic SNPs are present for distinct KIR family members in the NCBI Clinical Variations (ClinVar) database. These variations include malignant melanoma, posteriorly rotated ears, AIDS, delayed/rapid progression to (germ line), developmental delay and/or other significant developmental or morphological phenotypes, cleft upper lip, seizures, failure to thrive and abnormal facial shape (Supplemental Table S9). Genetic association with eQTL traits (insulin/insulin resistance, calcium, left ventricular hypertrophy, body fat distribution, body mass index and body weight, heart rate and heart failure, monocytes, glucose, diabetes Type 1 and schizophrenia) was seen for C1orf198, C2orf27, C16orf72, C21orf88 and GP2 (Supplemental Table S10).
Treatment of Ebola infections requires urgent attention if the current epidemic is to be contained. While vaccines and antibodies are being developed,18,49–51 alternative approaches based on novel molecular targets are urgently needed. Like any pathogen, the Ebola virus requires adsorption, entry and replication within the host cell. Thus, identifying the host proteins involved in EVD and understanding the pathways involved with these EVD-associated proteins is a first step toward rational host cell-based molecular targets discovery for therapy. The GWAS datasets provide a framework for identifying host genes relevant to pathogens, and numerous examples exist in the area of infectious diseases such as HIV, malaria and tuberculosis.15,17,18,52
The association evidence for the human proteins with the EVD and other diseases and disorders were obtained from four different bioinformatics tools at the level of : 1) transcriptome analysis using the NextBio Meta Analysis of the microarray datasets from the Array Express; 2) disease relationship using the MalaCards and the On Line Mendelian Inheritance in Man (OMIM) database, 3) pathways analysis using the pathways enrichment tools for the GeneALaCart and the DAVID functional Annotation tools and 4) nucleotide polymorphism, snp-genetic association using the Genetic Association Disease and the NCBI PheGeni association databases. Mining the disease-oriented databases generated identification of 45 human proteins associated with EVD. These proteins included the KIR family of natural killer cell receptors, chemokine (C-C motif) ligand 8, complement component 1, q subcomponent, B chain (C1QB), coagulation factor V (F5,) interferon regulatory proteins (IFIT2, IRF9), an FXYD domain containing ion transport regulator 3 (FXYD3) and several enzymes. In addition, 16 previously uncharacterized ORFs were also identified in this study.
Members of the killer cell immunoglobulin-like receptor family (KIR) offer an attractive target for EVD. The two activating KIR members, KIR2DS1 and KIR2DS3, are strongly associated with fatal outcome in the Zaire variant of EVD.48 It is suggested that the activation of these two KIR members may cause over-activation of the immune response, leading to rapid depletion of NK cells and lymphocytes. If so, it is tempting to speculate that inhibitors of these KIR family members might have direct or indirect therapeutic value.
In a recent study using whole genome transcriptome analysis involving Ebola virus-infected nonhuman primates, Yen et al.,18 identified a subset of differentially expressed genes including chemokine ligand 8 (CCL8), Complement Component 1, Q Subcomponent, B Chain, (C1QB), FXYD domain containing ion transport regulator 3 ((FXYD3), DEXH (Asp-Glu-X-His) box polypeptide 58 (DHX58), indoleamine 2,3-dioxygenase 1(IDO1) and platelet factor 4 variant 1 (PF4V1). These proteins, particularly the CCL8 chemokine, showed a strong correlation with survival.18 All of these proteins were identified in the current study using disease association approaches.
The 16 novel ORFs characterized in this study encompass enzymes, receptors, a transcription factor, a glucose suppressor and a secreted transmembrane factor. Using the mouse and hampster models53,54 the relevance of these 45 proteins to Ebola virus infection can be readily established using knockout technology such as antisense or siRNA. Since several of these proteins are readily detected in the body fluids, the efficacy of knockouts can be monitored with ease. These experiments would likely lead to verification of druggableness of some of the targets identified in the current study.
Mining of the drug databases identified numerous compounds including various FDA-approved drugs implicated in the pathways involved with the EVD-associated proteins. These drugs included amino acids, antineoplastics, nucleotide analogues, antibiotics, anti-inflammatory compounds, hormones, cytokines, metabolites and statins. Recently, FDA-approved selective estrogen receptor modulators were shown to inhibit Ebola virus infections.55 With the recent development of mouse and hamster models to study Ebola infection,53,54 the compounds implicated in this study can be rapidly validated for potential therapy use. These efforts can help to fill the gap in current treatment options and potentially expand the supportive care for EVD patients. An advantage of testing these compounds is that the majority of them have already been approved by the U.S. FDA. Hence, toxicology information already exists for these drugs.
The involvement of the EVD-associated human proteins with a complex landscape of Ebola-related and nonrelated diseases opens up novel opportunities for benefitting these diseases. Thus, if at least some of the 45 targets are verified for Ebola relevance, therapeutic approaches aimed towards the verified target could in theory expand the scope of potential usefulness across a spectrum of diseases.
This study identified 45 proteins including 16 previously uncharacterized ORF proteins present in the human proteome showing association with the Ebola virus disease and associated infectious and other diseases. Detection of these proteins in diverse body fluids and the identification of protein classes encompassing cytokines, enzymes, transporters and receptors provide a framework to explore both the diagnostic and drug therapy potential of these proteins.
The drugs (including those already FDA approved) implicated with the genes identified in this study provide a basis for their expanded use in supportive care for Ebola infections. This could fill in the interim needs until effective vaccines and other means of therapy are developed.
This work was supported in part by the Genomics of Cancer Fund, Florida Atlantic University Foundation. I thank Dr. Stein of the GeneCards team for generous permission to use the powerful GeneALaCart tool; Dr. Montague, Kolker Laboratory of the MOPED Team for batch analysis of the ORFs and the Human Proteome Map and Proteomics DB for the datasets. I thank Jeanine Narayanan for editorial assistance.
The author declares no conflict of interest.

©2014 Narayanan. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.