MOJ
eISSN: 2373-4442


Research Article Volume 1 Issue 4
athology Department, Salmaniya Medical Complex, Bahrain
Correspondence: Mohamed Jahromi, Mohamed Jahromi, Translational Research Group, Clinical Research Unit, Directorate of Clinical Services, Dasman Diabetes Institute, P.O Box 1180, Dasman 15462, Kuwait,
Received: May 30, 2014 | Published: October 31, 2014
Citation: Jahromi M, Ahmed A, Behbehani K (2014) Networking Between Cytokine Gene Polymorphisms Profile and HLA in the Pathogenesis of Type 1 Diabetes (A Pilot Study). MOJ Immunol 1(4): 00023. DOI: 10.15406/moji.2014.01.00023
Type 1 Diabetes (T1D) is an autoimmune disease with both genetic and environmental components. More than 60 genes have been identified to affect the risk of T1D, with the HLA loci having the greatest impact on susceptibility. The association of T1D with alleles at HLA loci, especially the HLA class II genes DR and DQ, is well-established. However, other candidate genes may also have some significant impact on genetic susceptibility.
Cytokines are central mediators of inflammation by controlling innate and adaptive immune responses as well as tissue damage, defense, repair, and remodeling. T1D is an inflammatory disease of the pancreatic islet, in which insulin-producing β-cells are preferentially destroyed to varying degrees by the concerted action of auto reactive T-cells. A number of cytokines have been shown to be important for the development of T1D both at the level of the immune system and at the level of the target β-cells. The actual mechanism of β-cell destruction is still unclear, however, according to the available cytokine genetic studies, β-cell destruction might be predicted, Figure 1.
As an autoimmune disease that starts with inflammation of the pancreatic β- cells leading to β- cell death, cytokines may play a significant role in the overall pathogenesis of T1D. To determine the effects of cytokine gene polymorphisms in the pathogenesis of T1D, high risk interferon-gamma (IFN-γ), interleukin-6 (IL-6) as well as transforming growth factor β (TGF-β) gene polymorphisms were considered in HLA DRDQ fixed patients with T1D.
In this pilot study, we determined that there may be critical networking among different cytokines and HLA which may play a role in the pathogenesis of T1D. However, a larger scale case control study is required to confirm these findings and to determine the functionality of cytokines and the high-risk HLA patients.
Keywords: haplotype, type 1 diabetes, gene to gene networking, cytokine gene polymorphism, HLA
T1D, type 1 diabetes; IDF, international diabetes federation; GWA, genome-wide association
The prevalence of diabetes is increasing linearly and its burden is among the fastest growing diseases globally.1,2 International Diabetes Federation (IDF) estimates that more than 382 million people worldwide have diabetes and that this number is likely to reach 592 million by 2035. T1D which predominantly affects young people is increasing at an alarming rate of 3% per year. It has been estimated that around 78000 children ≤ 14 years of age develop T1D annually.3
The destruction of insulin-producing pancreatic β-cells by a beta-cell specific autoimmune process is considered to be the main cause of T1D. There are many factors that may directly or indirectly trigger the onset of T1D;1,2 among these, genetic as well as environmental factors play a dominant role in determining the pathogenesis of T1D.1,4 Although HLA acts as the determinant genetic susceptibility factor for the onset of T1D; HLA-mediated susceptibility represents only ∼50% of the genetic susceptibility; therefore, HLA is not an adequate measure to evaluate the risk of T1D.4,5
There are compiling reports associating other candidate genes within or linked to the MHC and non-MHC genes with T1D.4-7 These studies indicate critical and significant interactions among different genes and/or loci within or linked to the MHC or non-MHC genes.7
A number of cytokine gene polymorphisms have been shown to be important for the development of T1D both at the level of the immune system and at the level of the target β-cells.8-17,20 The actual mechanism of β-cell destruction is still unclear, however, according to the available cytokine genetic studies, β-cell destruction might be predicted, Figure 1.8-14,18-20.
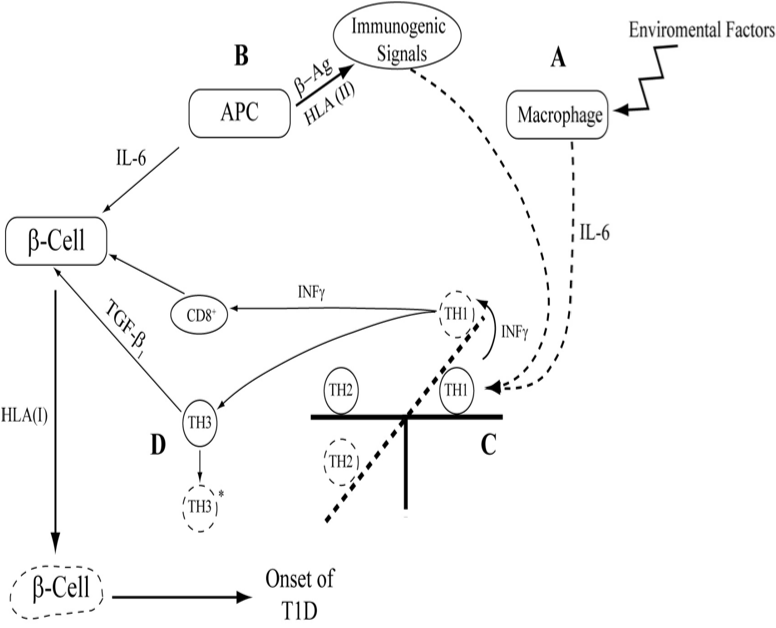
Figure 1 This schematic diagram explains the possible effects of cytokines in the initiation triggering of T1D by signals from either environmental factors “A” or immunogenic factors “B” or both Release of IL-6 would change the counterbalance between TH1/TH2 “C” cytokines level. Increase level of TH1, IFN-γ, cause the cytotoxic process of breakdown of β-cell by CD8. As a result of deviation of cytokine cross regulation to TH1 dominance TH3 or T reg, regulatory T cell lymphocyte subset produce FoxP3 and immunosuppressive cytokines, TGF- β1, however, due to genetic malfunction this cytokines are proved not to be able to bring TH1/TH2 back to normal position, D. This will lead to the death of pancreatic β cell. This is a simplified schematic description of the impact of cytokines in the pathogenesis of T1D.
Few inflammatory proteins have been demonstrated to be critical for T1D development in vivo.17,18 The genetic susceptibility of polymorphisms in Interferon-gamma (IFN-γ) CA-repeat, Interleukin-6 (IL-6 G (-174) C in the promoter region,9 Transforming Growth factor- β1 (TGFβ1T(29))C in codon 1010 are well established.21-24
Knowledge of the genetic architecture of T1D has increased recently owing to large-scale genome-wide association (GWA) studies.24-27 Estimates of the contributions of the HLA regionand numerous non-HLA loci across the genome now account for the understanding of the pathogenesis of the disease. Genetic risk for T1D is likely to be due to interactions between several susceptibility genes or loci in the same biochemical pathway.28 Due to the elemental role of cytokines in inflammation and autoimmune onset of T1D, this study was performed to investigate the possible gene to gene interactions between the reported high risk cytokine gene polymorphisms 8-10 and susceptible HLA class II alleles in the pathogenesis of T1D.
In this study, we enrolled 182 Caucasian patients with T1D who were studied for cytokine gene polymorphisms.8-10,28 This work was carried out upon receiving the Institution’s ethical approval. Patients were segregated manually according to their HLA-DR3/4/DQ8 haplotype. The case group consisted of 98 patients with the same DR/DQ, and the control group consisted of 84 patients with HLA haplotypes other than DR3/4/DQ8.
Evaluating risk of developing T1D depends on determining an individual’s HLA type, especially of the HLA DRB1 and DQB1 alleles. Individuals positive for HLA-DRB1*03 (DR3) and/or HLA-DRB1*04 (DR4) with DQB1*0302 (DQ8) have the highest risk of developing T1D.6
Cytokine gene polymorphism results for both case and control groups were typed accordingly. The frequency of matching genotypes of cytokine gene SNPs in both case and control groups were determined using Fisher’s Exact Test.
Patients were segregated according to their HLA haplotypes, as previously reported.7 In order to examine the effect of cytokines, other than the well-established HLA effects, in genetic susceptibility, patients with HLA DR*030/DR*040/ DQ*0302 who met high risk cytokine genotypes were segregated as case group. In other words, by fixing our patients for well-established high risk HLA haplotypes (DR3/DR/4/DQ8) only patients with high risk cytokine genotypes were considered as the positive case for gene to gene networking. All other HLA non- (DR3/DR/4/DQ8) were considered as the control group.
Patients were segregated according to their HLA DR/DQ haplotypes along with their associated reported high risk cytokine genes. Interestingly, 70 (71%) of cases (HLADR3/4DQ8) had similar reported high risk cytokine polymorphism genotypes (IFN-γ (122/121)/IL-6 (G/G)/TGF-β (C/T))9,10 compared to 28 (33%) in control group (Non DR3/4/DQ8) who had none of the cytokines high risk gene, p=<1 , Odd Ratio=5.0 with 95 % confidence interval = 2.7-9.4 (Table 1).
Haplotypes |
+ve |
-ve |
p value |
Odd Ratio |
95 % CI |
HLA-DR3/4/DQ8/122/121 / G/G /C/T (cases) |
70 |
28 |
< 0.0001 |
5.00 |
2.7-94 |
Non (HLA-DR3/4/DQ8/122/121 / G/G /C/T) (Controls) |
28 |
56 |
Table 1 Patients with type 1 diabetes were segregated according to their DR*0401/DR*0301/DQ*0302. Of nighty eight patients with high risk HLA 70 (71%) had high risk gene for studied cytokines in comparison. IFNG CA repeat 122/121, IL-6 (-174) G/G and TGFB (29) C/T were considered as the high risk as reported [9-11]. Majority of patients with high risk HLA DR3/4/DQ8 had had high risk genotype for IFNG, IL-6 as well as TGFB p=<0.0001, Odd Ratio=5.0 and 95 % CI=2.7-9.8.
The incidence of T1D is increasing at an alarming rate and is reaching above that which is predicted by the IDF. HLA accounts for almost 50 % of genetic susceptibility to T1D. According to the recent GWAS, more than 60 non-MHC genes or loci are associated with susceptibility to T1D. Genetic networking stands for interactions between the associated genes or loci with T1D regardless of their genetic susceptibility share. Although HLA, insulin gene and PTPN22 indicate higher susceptibility to T1D,29 other genes and loci seem to have a critical and significant impact in the pathogenesis of T1D.30,31
This critically significant association of high risk HLA patients with high risk cytokine gene polymorphisms emphasize on the phenomenon of gene to gene networking in the pathogenesis of T1D. Considering the role of cytokines in the mediation of autoimmunity and inflammation, the current finding is valuable, however, further investigations with larger population studies is required.
We would like to acknowledge Professor John Todd, Cambridge University for providing full HLA data for Warren Families and his fruitful discussion. Authors would like to thank Dr. Anwar Mohammad for his time and help in the figure 1 and appreciate. Dr. Amal Hassan for her time and proof reading the article.
This is to confer that the author has no any conflicts of interest or received any financial support for the presented manuscript.

©2014 Jahromi, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.