MOJ
eISSN: 2573-2919


Research Article Volume 6 Issue 4
1São Paulo State University, Technology Department, Jaboticabal, São Paulo, Brazil
2Federal University of Mato Grosso do Sul, Chapadão do Sul, Mato Grosso do Sul, Brazil
3National Laboratory of Scientific Computing, Petrópolis, Rio de Janeiro, Brazil
4Center for Nuclear Energy in Agriculture, University of São Paulo, Piracicaba, São Paulo, Brazil
Correspondence: Acacio Aparecido Navarrete, Federal University of Mato Grosso do Sul, Rod. MS-306, km 105, Chapadão do Sul, MS, Brazil, Zip Code 79560-000, Tel +55-67-3562-6352
Received: June 24, 2021 | Published: July 5, 2021
Citation: Pedrinho EAN, Navarrete AA, Kishi LT, et al. Taxonomic and nitrogen-cycling microbial community functional profiles of sugarcane and adjacent forest soils in Southeast Brazil. MOJ Eco Environ Sci. 2021;6(4):119-125. DOI: 10.15406/mojes.2021.06.00224
Nowadays, due to the expansion of agricultural borders, it is highly desirable to increase the sustained productivity of sugarcane cultivars using the knowledge of soil microbial communities. In this study, twelve shotgun metagenomic datasets based on genomic DNA from soil were analyzed using the Metagenomics Rapid Annotation using Subsystem Technology (MG-RAST) and Statistical Analysis of Metagenomic Profiles (STAMP) to assess differential responses for the total soil bacterial community composition and nitrogen-cycling microbial community functional potential in soils from sugarcane field with pre-harvest burning and adjacent forest in dry and wet seasons in Southeast Brazil. The soil bacterial community revealed higher abundance for Actinobacteria in forest soil than sugarcane soil in dry and wet seasons, and an opposite pattern for Proteobacteria and Planctomycetes in these soils in both seasons. The results obtained in this study based on the KEEG map suggest that the forest soil has a higher nitrogen-cycling microbial community functional potential compared to the sugarcane soil, independently of the season. The gene sequences associated with carbohydrate metabolism were the most frequent in all soil metagenomes. Taken together, the results confirm previous findings regarding the effects of forest conversion to sugarcane production area, providing new insights regarding to this conversion through the prism of the seasonality and pre-harvesting method on microbially mediated nitrogen cycle in sugarcane production fields.
Keywords: soil bacterial community composition, shotgun metagenomic sequencing, nitrogen cycle, seasonality, sustainability
Brazil is a major sugarcane producer and Southeast region cultivates 5.3 million hectares, close to 50% of Brazil’s sugarcane area.1 The rapid increase in production has brought into question the sustainability of biofuels, especially considering the sustainable use of the soil associated to its ecosystem services. The continuous cropping of sugarcane can results in the disruption of the soil microbial community and cause significant declines in yields.2 Pre-harvesting burning of sugarcane straw is an agricultural practice that, despite the state legislation recommend it to be completely phased out in areas where mechanized harvesting is possible, it is usual in non-mechanized areas.3 Despite the previous studies focusing the soil microbiota in sugarcane burnt soils,4–7 there are a lack of conclusive data about the impact of this harvest method on the tropical soil ecosystem sustainability, mainly regarding to the microbe-mediated processes in soil.8 Determination of the effect of agricultural management practices on soil sustainability is a challenge, particularly the aspect concerning the evaluation of the biological quality of the soil.9
A widely accepted definition of soil quality is the capacity of a soil to function within ecosystems and land-use boundaries to sustain biological productivity, to maintain environmental quality, and to promote plant, animal, and human health.10 Taxonomic and functional changes in the microbial community of soil can reflect changes in the soil quality.11 Soil microorganisms are involved in ecological functions that are essential to the environment, such as decomposition of plant and animal residues, solubility of nutrients, formation of organic matter, soil structure, plant growth promoters, and biological control agents. Thus, microbial processes are fundamentally important to the functioning of agro-ecological production systems, which perform functions directly related to their productivity and sustainability.12
The shotgun metagenomic sequencing and bioinformatic analysis are important tools to quantify the abundance and distribution of microbial taxa at the community ecological level, gene families involved in soil microbial functions and nutrient metabolism below-ground.13 The size, composition, and function of these microbial communities are affected by land-use,14,15 sazonality,16 agricultural practices,17,18 and others environmental changes. De Caceres, Legendre, and Moretti19 consider that differential abundances in microbial taxa and functional genes can be used as potential bioindicators for environmental change. However, our knowledge on the soil microbial communities, and below-ground nitrogen metabolism in tropical soils under agricultural and forestry land-uses is limited and requires further elucidation of this ecological aspects related to soil quality concerning to vegetation cover, sazonality and agricultural practices.
In this study, we evaluated the seasonal patterns in taxonomic and functional profiles of the soil microbial communities in sugarcane and adjacent native forest areas located in the Southeast Brazil. We evaluated the following hypothesis: (1) forest soil presents higher abundance of the most bacterial phyla in wet and dry seasons in comparison with sugarcane soil, and (2) higher nitrogen-cycling microbial community functional potential is found in forest soil than in sugarcane soil in wet and dry seasons. To evaluate these expectations, we used whole genome shotgun metagenomic sequencing of DNA obtained from soil samples to generate raw data for subsequent bacterial profiling and functional analyses.
Sampling sites and soil sample collection
Two different field sites were chosen in Southeastern Brazil, a burnt sugarcane (SUG) harvesting site located in a private property (Fazenda Coqueiro) (21˚42'30.67"S 47˚34'56.89"W) and a native forest (FOR) site located in the Vassununga State Park (21˚42'34.62"S 47˚34'30.04"W). Both the sites are located in Santa Rita do Passa Quatro municipality in the state of São Paulo, Brazil. The climate in the region is classified as Cwa (Koppen’s classification), with an annual precipitation varying from 1,300 to 1,700mm, and total precipitation of 18.6mm in the driest month (August). The average annual temperature in the region is 23.3˚C, with an average maximum temperature (December to February) of 26˚C and a minimum of 19.50˚C (June to August).
Soil samples were collected from three different points at least 14m apart from each other along a transect at each sampling site in both the dry (D) (February 2011) and wet (W) (November 2011) seasons. Undeformed soil cores were taken from the 0- to 20-cm top-soil layer after the removal of the litter layer using a 5-cm diameter aseptic cylindrical corer. A total of 12 soil samples were collected from the field (two sampling sites×three sampling points×two sampling periods). The samples were transported to the laboratory in a cool box containing ice, stored at −20˚C, and processed within 72h of sampling.
Isolation of DNA from soil and high-throughput sequencing of soil metagenomes
Soil DNA was extracted from 0.5g (wet weight) of 24 soil samples using the FastDNA™ SPIN Kit for Soil (MP Biomedicals, Santa Ana, CA, USA) and the FastPrep® equipment (MP Biomedicals, Santa Ana, CA, USA) according to the manufacturer’s instructions. Both purity and quality of the genomic DNA were assessed via spectrophotometry on a NanoDrop apparatus (NanoDrop ND-1000 NanoDrop Technologies, Inc., Wilmington, DE, USA) to determine the absorbance at the following wavelengths: 230, 260, 280, and 320nm. The DNA concentration was determined using a Qubit 2.0 fluorometer (Life technologies, Carlsbad, CA, USA). DNA was stored at −20˚C until further use.
Twelve DNA sequencing libraries (three FOR-D libraries, three FOR-W libraries, three SUG-D libraries, and three SUG-W libraries) were prepared using the Illumina Nextera sample preparation kit (Illumina, San Diego, CA, USA) according to the manufacturer’s instructions. The libraries were evaluated on the 2100 Bioanalyzer using a High Sensitivity DNA kit (Agilent, Santa Clara, CA, USA) to estimate the library size. The libraries were quantified using the Qubit dsDNA HS kit on a Qubit 2.0 fluorometer (Life Technologies, Carlsbad, CA, USA) and quantitative PCR (qPCR) was performed using the KAPA SYBR FAST qPCR Master mix, Illumina standards, and primer premix (KAPA Biosystems, Wilmington, MA, USA) according to the instructions provided by Illumina. The resulting DNA libraries were denatured with NaOH, diluted to 8pM using Illumina’s HT1 buffer, and spiked with 1% PhiX. Equal concentrations of the libraries were loaded on the Illumina’s HiScanSQ System (Illumina, San Diego, CA, USA) operated in a Rapid Run Mode to generate 2×100bp paired-end reads. After sequencing, the libraries were separated, and the BCL files were converted into FASTQ format using the Casava 1.8.3 software (Illumina, San Diego, CA, USA). The raw data of the sequencing were deposited in the National Center for Biotechnology Information (NCBI) server (https://www.ncbi.nlm.nih.gov) under accession number PRJNA665014 ‘Taxonomic and functional microbial profiles of sugarcane and forest soils in the tropical region’.
Shotgun metagenomic data processing and taxonomic identification of bacterial groups
The sequences of each of the twelve libraries were analyzed with the CLC Genomics Workbench 6.0.1 software, removing reads smaller than 35 bases, adapters and bases with less than quality phred 20. Taxonomy was assigned using the lowest common ancestor (LCA) in the MG-RAST server, with the parameters 1e05 as the maximum e-value, a minimum identity of 60%, and a minimum alignment length of 15 as the cutoff.
Statistical analysis was conducted using the Statistical Analysis of Metagenomic Profiles (STAMP).20 The significance of the relative proportion difference in the taxonomic distribution of metagenomic samples was performed using the two-sided Fisher's exact test, with the Newcombe–Wilson confidence interval method. Since the P-values were not uniformly distributed using the Storey's false discovery rate (FDR), the Benjamini–Hochberg FDR was used for the correction.
Nitrogen-associated genes and -metabolic pathway analysis
Nitrogen-associated cycle genes were identified using the SEED subsystems annotation source of MG-RAST, with a maximum e-value of 1e05, minimum identity of 60%, and minimum alignment length of 15. Subsets of the nitrogen-associated cycle genes in a functional subsystem for sequences belonging to Bacteria domain were generated using the workbench tool from the MG-RAST server. The nitrogen metabolic pathway analysis was performed using KEGG mapper from the MG-RAST server, with a maximum e-value of 1e05, minimum identity of 60%, and minimal alignment length of 15.
A functional analysis using the SEED21 and KEGG22 databases was performed using the STAMP software. Each sequence was related to its SEED functional role using the best BLAST score to protein sequences without known functional roles. A similar procedure was used to match each sequence to a KEGG orthology (KO) accession number.
Metagenomic profiles
The twelve libraries yielded over 15,000 reads. The readings identified as artificial duplicates were removed using MG-RAST. After quality control, an average of 76,607, 74,426, 63,005, and 166,676 proteins were predicted for FOR-D, FOR-W, SUG-W, and SUG-D, respectively, and 71.7%, 73.9%, 73.1%, and 78.5% of the total reads were annotated as functionally assigned proteins. The taxa richness estimated for FOR-D, FOR-W, SUG-W, and SUG-D revealed the following values: 284.599, 252.993, 381.042, and 385.731, respectively.
Bacterial community composition in sugarcane and forest soils in both dry and wet seasons
Proteobacteria, Acidobacteria, and Actinobacteria were the most abundant bacterial phyla found in all soil metagenomes (Figure 1). Higher abundance was showed for Actinobacteria in FOR-D and FOR-W than SUG-D and SUG-W, and an opposite pattern was revealed for Proteobacteria in these soils in both seasons (Figure 2a and 2b). These results corroborate findings of previous studies, which also reported that these phyla were the most abundant in soil samples.23–28 The predominance of Proteobacteria in such environments can be explained by the fact that this phylum comprises the most phylogenetically diverse group in the Bacteria domain, known to be metabolically versatile, including aerobic and facultative bactéria.29

Figure 1 Relative abundance of the most abundant bacterial phyla as revealed by metagenomic sequencing and measured by percentage of total Bacteria.
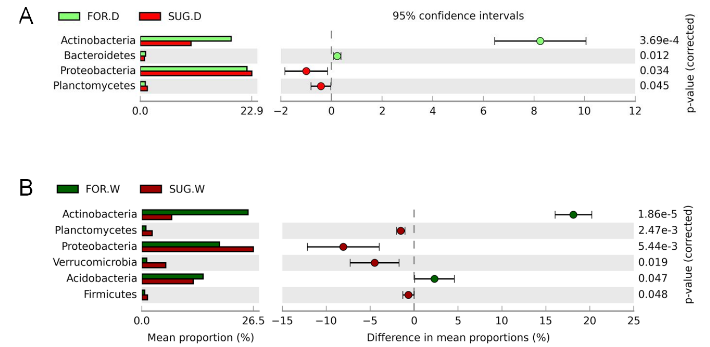
Figure 2 Differences in abundance of bacterial phyla revealed using the Statistical Analysis of Metagenomic Profiles (STAMP) software based on abundance of metagenomic sequences. (a) Differences between soil samples from native forest (FOR-D) and sugarcane (SUG-D) both collected during the dry season. (b) Differences between soil samples from native forest (FOR-W) and sugarcane (SUG-W) both collected during the wet season.
The prevalence of Acidobacteria ranged from 2.4% to 78.5% in different soil samples. Acidobacteria form one of the main groups found in tropical soils, with prevalence in tropical forest soil.14,17 This corroborates the results found in FOR-D (26.8%) and FOR-W (26.2%), in contrast to the SUG-W sample, where only 13.5% of the sequences corresponded to the phylum Acidobacteria. According to Ward et al.30 and de Chaves et al.,13 the occurrence of Acidobacteria in soil may indicate the presence of genes related to the synthesis of cellulose and proteins, and carbon degradation, suggesting characteristics of resistance to desiccation and metabolic potential for C degradation in sugarcane soil.
The relative abundance of Actinobacteria was lower in sugarcane soil, SUG-D (11.8%) and SUG-W (5.9%), while it was higher in native forest soils, FOR-D (18.3%) and FOR-W (27.1%) (Figure 2a and 2b). The data also showed that Actinobacteria was the dominant phylum in the native forest samples in dry and wet seasons. Russo et al.31 have reported that Actinobacteria is a dominant class in sandy soil, and moisture is a determining factor for abundance of this phylum, which increases in arid soil due to the resistance of several species to drought stress.32,33
Other bacterial phyla with differential responses in abundance regarding to sugarcane and forest soils in the dry and wet seasons were Bacteroidetes, Planctomycetes, Verrucomicrobia, and Firmicutes (Figure 2a and 2b). Verrucomicrobia was more abundant in SUG-W (5.98%) than in SUG-D (1.30%), FOR-D (1.34%), and FOR-W (1.04%) samples. The comparison of the abundance of bacterial phyla in FOR-D and SUG-D samples showed significant differences for Acidobacteria, Bacteroidetes, Proteobacteria, and Planctomycetes (Figure 2a), whereas the comparison of FOR-W and FOR-W samples revealed significant differences for Actinobacteria, Planctomycetes, Proteobacteria, Verrucomicrobia, Acidobacteria, and Firmicutes (Figure 2b).
Overall, these results reveal a differential distribution of bacterial taxa across the soil profile in soils from sugarcane field with pre-harvest burning and adjacent forest in dry and wet seasons in Southeast Brazil, based on possibly the available nutritive resources in these soils in the dry and wet seasons. Some microbial communities adapt easily to the accessible carbon substrates in the native forest soils, while others adapt to recalcitrant carbon substrates and inorganic nutrients in the sugarcane soil.13,15 However, these bacterial taxa can be the main contributors to soil ecosystem functions because they are particularly abundant in this habitat. These differences may be associated with the soil characteristics such as nutrient availability, moisture, and texture, which are also determinants to the microbially-mediated biogeochemical processes.16
Functional genes associated with nitrogen metabolism in sugarcane and forest soils
Functional analysis of the soil metagenomes showed significant differences between the SUG-W and SUG-D samples and the FOR-D and FOR-W samples regarding to the abundance of nitrogen-associated genes and -metabolic pathway (Figures 3–5). The resulting functional subsystems may represent various metabolic functions found in the sugarcane and forest soils studied in the dry and wet seasons. The carbohydrate subsystems were highlighted at level 1 of the functional subsystems with significant differences for FOR-D and SUG-D samples in the dry season (Figure 3) (Figure 4). This group includes the enzymes involved in the tricarboxylic acid (TCA) cycle and pathways for the degradation of glucose to pyruvate (i.e., the Embden–Meyerhof, Entner–Doudoroff, and pentose pathways). The second most prominent category among the metagenomes was miscellaneous (FOR-D), fatty acids, lipids, isoprenoids (FOR-W and SUG-D), and RNA metabolism (SUG-W). The subsystems of amino acid metabolism, carbohydrates, cofactors and vitamins, and energy metabolism showed a similar profile for all metagenomic samples according to the MG-RAST and SEED annotation sources.

Figure 3 Nitrogen metabolic pathway analysis performed using KEGG mapper (http://www.genome.jp/kegg/mapper). The red box indicates the enzyme that had a significantly high relative abundance in native forest site sampled in the wet season. The blue box indicates the enzyme that had a significantly high relative abundance in native forest site sampled in the dry season.

Figure 4 Nitrogen metabolic pathway analysis performed using KEGG mapper (http://www.genome.jp/kegg/mapper). The red box indicates the enzyme that had a significantly high relative abundance in sugarcane site sampled in the dry season. The blue box indicates the enzyme that had a significantly high relative abundance in sugarcane site sampled in the wet season.
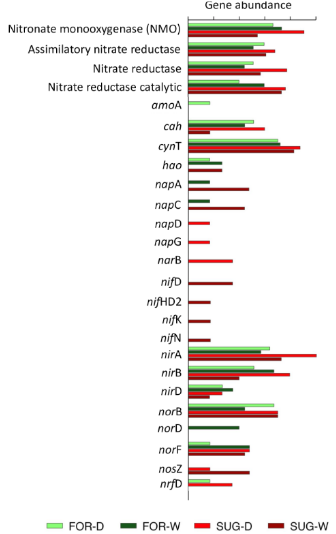
Figure 5 Absolute sequence abundance of microbial functional genes associated with nitrogen metabolism based on metagenomic sequencing data for each of three libraries per sampling point in native forest and sugarcane sites in both dry and wet seasons.
Similar results obtained from the functional categories of the SEED Subsystems and KEGG showed that the genes involved in the metabolism of amino acids, carbohydrates, and energy were common among the extensively studied metagenomes (Figure 5). The comparison of the ratio of metabolic categories based on the SEED and KEGG database showed that the metabolism of carbohydrates was dominant among the metagenomic samples, especially the dry season samples (FOR-D and SUG-D) (Figures 3–5).
The SEED and KEGG classification results showed high abundance of genes involved in energy metabolism. These genes were classified as participants of nitrogen metabolism (Figure 3) (Figure 4). Thus, the functional analyses of the metagenomic datasets demonstrated that the key genes involved in the N metabolism were related to the energy cycles. For example, in the subcategory of oxidative phosphorylation, most of the sequences showed similarity to enzymes related to energy storage (respiratory chain) such as coxA (cytochrome C oxidase subunit I), PPA (inorganic pyrophosphatase), NDH (NADH dehydrogenase), NADH quinone N oxidoreductase subunits L, M, F, H, and D. Additionally, the following sequences associated with enzymatic functions were also observed: methane metabolism (carbon), enzymes that catalyze oxidation-reduction reactions (cutL/coxL, coxS, and cutM/coxM), sulfur (sulfate adenylyltransferase), nitrogen (nirA-ferredoxin-reductase nitrite, nirB-nitrite reductase large subunit, and nitronate monooxygenase), and carbon sequestration. However, the enzymes involved in the synthesis of carbohydrates by photosynthesis (fructose-6-phosphate phosphoketolase) were lower than those involved in other metabolic pathways.
The results obtained in this study based on the KEEG map suggest that the FOR-D and FOR-W samples have a higher nitrogen-cycling microbial community functional potential due to the higher abundance of genes involved in nitrogen metabolism compared to that of SUG-W and SUG-D samples. Assuming that the functional structure of the community changes with land use conversion, similar to the results observed for the bacterial community composition, this fact justifies the changes in the C and N microbially-mediated processes in the soil.
Comparative metagenomic analyses have indicated that the substrate (i.e., soil or water) plays a fundamental role in determination of the taxonomic and functional profiles of microbial communities. Therefore, soil samples tend to be more similar to each other with respect to taxonomy and the presence of environment-specific genes than the similarity observed in samples from other environments.34 In FOR-D and FOR-W, nutrient contents and vegetation may be the most important factors affecting the soil bacterial community, while in SUG-D and SUG-W, in addition to these factors, stress conditions (caused by temperature, UV, and drought) might also affect the microbial functionality in soil.
Among the genes restricted to the denitrification process, we found genes belonging to groups norB, norC, norD, and norF involved in the reduction of nitric oxide; however, only norB and norF were present in all the analyzed metagenomes. The presence of groups of genes that encode nitrate reductase, which are responsible for the reduction of nitrate to nitrite, was not found except for naiD in FOR-W. The nitrite reductase group of genes, nirA, nirB, and nirD, which are responsible for reducing nitrite to ammonia, were present in all the metagenomes and with the highest prevalence in the metagenome FOR-D. This occurrence of genes involved in nitrite and ammonia assimilation suggests the retention of N and consequently contribute with lower loss by denitrification.
The genes nifH, nifK, nifN, and nifW, which encode the nitrogenase reductase enzymes, and the gene nifB, which encodes the nitrogen binding protein, were found only in the SUG-D metagenome, probably because the lack of these genes in other metagenomes is due to the low copy numbers which compromises their identification (Figure 5). Another subunit of the nif gene (nifD) was identified in the SUG-D and SUG-W metagenomic samples, possibly due to soil handling.
The presence of these functional genes associated to N fixation in the metagenomes of soils from sugarcane field with pre-harvest burning and adjacent forest in dry and wet seasons can be explained by the abundance of organisms belonging to the Rhizobiales order (data not shown), as determined by Ribosomal Database Project (RDP) library compare tool using Naive Bayesian classifier.35 The Bradyrhizobium genus has an important role in N and C fixation and biodegradation of aromatic compounds.36
Taken together, the results of the current study showed higher abundance of the most bacterial phyla and nitrogen-cycling microbial community functional potential in wet and dry seasons in forest soil in comparison with sugarcane soil. Hence, both hypothesis of this study are acceptable based on these results. The differential distribution of bacterial taxa across the soil profile in sugarcane field with pre-harvest burning and adjacent forest site in dry and wet seasons indicates that land use, harvesting management and seasonality is having an effect on soil bacterial community composition affecting the integration of carbon and nitrogen pathways related to the energy metabolism. These findings therefore tighten the importance of policies for extinguishing pre-harvest sugarcane straw burning in an effort to improve sugarcane ethanol sustainability based on the nitrogen-cycling microbial community functional potential.
We thank Gabriel Henrique Sant’Ana Pereira, manager of Vassununga State Park, for support in field work.
This work was supported by Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP, Proc. 2009/54274-9), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior-Brasil (CAPES) — Finance Code 001 and 23038.006927/2014-35/Premium 116/2017. E.G.M.L. Thanks CNPq for the Research Productivity Grant (305032/2019-4).
The authors declare that there is no conflict of interest.

©2021 Pedrinho, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.