MOJ
eISSN: 2374-6912


Research Article Volume 2 Issue 4
University of Hull, Saudi Arabia
Correspondence: Bassam Niaz, University of Hull, Saudi Arabia, Tel 00966506994412
Received: February 15, 2015 | Published: September 7, 2015
Citation: Niaz B. Study of velocity and chemotaxis toward l-serine in normal and filamentous forms of escherichia coli. MOJ Cell Sci Rep. 2015;2(4):93-102. DOI: 10.15406/mojcsr.2015.02.00034
This study is concerned with the measurement of velocity of normal and filamentous cells of E. Coli in the presence of l-serine (a Chemoattractant). Therefore, experiments with many steps were applied to find the effect of cell size on its swimming velocity under chemotactic stimulation. A dunn chamber was used to measure the swimming velocity of motile cells and filaments toward the attractant. Filament formation was stimulated by the addition of cephalexin (an b-lactam antibiotic). Cell division was blocked and cell growth continued for several hours. Although they have an elongated body, filaments exhibited motility and chemotaxis in a similar manner to normal cells of E. Coli. The behaviour however was different in the absence or presence of a Chemoattractant. Normal cells ran and tumbled alternately, whereas, filaments ran and stopped alternately in the absence of a Chemoattractant. In the presence of a Chemoattractant, both kinds of cells ran continuously, followed by adaptation mechanism. Filaments were found to be faster than normal cells and the suggested explanation to the increased velocity of filaments over time is due to:
Troubleshooting, Pros And Cons Of Dunn Chamber Are Discussed As It Was Used For Chemotactic Study Of Free-Swimming Organisms For The First Time. Furthermore, The E. Coli Chemotaxis System Is Explained.
Keywords: escherichia coli, e. Coli, filament, velocity, chemotaxis, chemoreceptor, dunn chamber, gradient
Escherichia coli
Escherichia coli are the most well studied motile bacteria.1 They are rod shaped, motile, enteric (intestinal), facultative anaerobic bacteria and their optimal growth is at 37°C (98.6°F). The approximate size of E. coli is ≈1µm in diameter and ≈2µm long. They move by using about six flagella, which are positioned randomly on their surfaces and are used for locomotion. Rotary motors at their bases power these flagella, and each flagellum is 10μm long and 20nm wide.2–4 The motivation for movement in E. coli is due to two important reasons: finding food and escaping from poisons.5 For instance, E. coli swim toward sugars (galactose, glucose, ribose and maltose),6 amino acids (aspartic acid and serine),7 electron acceptors (nitrate and oxygen),8 Pyrimidine and dipeptides.9 On the other hand, they swim away from potentially noxious chemicals such as fatty acids and alcohol.10,11
Flagellar rotation in Coli cells
There are two types of flagellar rotation in E. coli; firstly, the flagellar motors turn counter clockwise (CCW) by forming a pack together at one side that drives the bacterium forward (also called “run”) and normally lasts nearly 1 to 2seconds. Secondly, the flagellar motors turn clockwise (CW) by pulling in opposite direction causing a bacterial “tumble” motion and normally takes less than a second,as shown in Figure 1.12,13
In a normal situation (lack of stimuli), runs and tumbles occur in an alternating arrangement, with every run creating a three-dimensional step in random movement.14 In normal cells of E. coli, the swimming velocity is about 20μm/s and the rotational velocity is about 10Hz.15
Chemotaxis
In the 1880s, the first discovery of chemotaxis in bacteria by Engelmann10 and Pfeffer11 took place. They found that some compounds attract the bacteria and others repel them. In their experiments, they observed the bacteria congregating around oxygen-producing chloroplasts and escaping from noxious acids.16 However, the precise definition of chemotaxis process is the movement of the organisms or cells toward or away from certain chemicals.8
If the swimming condition is suitable in the presence of a spatial gradient of a certain attractant, a bias on the random movement that drives the bacteria in a favourable direction will be forced, as shown in Figure 2.17 When the bacteria are faced with an attractant, tumbling is repressed and running is stimulated, which results in movement toward the attractant. When the bacteria are faced with a repellent, tumbling is promoted and running is repressed, which stops their movement toward the repellent.18

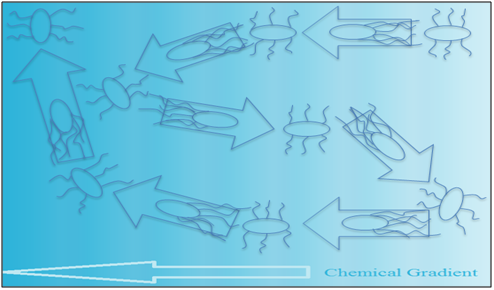
coli chemotaxis system E. Coli chemo-sensors: Sequencing of Escherichia coli genome reveals it has chemotaxis sensors. Ribose and galactose sensed by Trg,19 maltose and aspartate by Tar,20 serine by Tsr,21 dipeptides by Tap,22 and the recently discovered Aer,23 which might be a redox detector. Vast arrays of internal and external signals are monitored by the chemotaxis signal transduction system of E. coli. Five membrane sensors and the phosphotransferase system of cells perceive extracellular nutrients, intracellular redox potential, pH, and temperature of repellents.24
Information obtained from different parts of the chemotaxis system is often contradicting and therefore it must be integrated to give one motor response. The chemotaxis signal transduction system processes sensory information at all levels. Tar and Tsr are the major sensors in E. Coli, which have their independent but parallel processing systems. Minor receptors connect to these processing systems.24
Coli chemoreceptors: Bacteria sense chemical stimuli in the environment by series of chemoreceptors, which are made of transmembrane proteins, the methyl-accepting chemotaxis proteins (MCPs). The chemoreceptors conduct the sensation to a signal transduction system located in the cytoplasm. The system is made up of two compartments, the kinase (CheA) and the response regulator (CheY).24
Chemoreceptors, in the presence of repellents in the surrounding environment, activate the CheA kinase, which in turn increases CheY response regulator phosphorylation process. As a result of phosphorylation, the response regulator changes its conformation leading to its attachment to the flagellar motor’s switching proteins; this binding causes the repellent response. Attractants on the other hand, work in the opposite way by inhibiting the phosphorylation process at the response regulator and as a result the connection between the response regulator and flagellar motor is cut. Cells of E. coli do not respond to absolute levels of repellents or attractants, but rather to the changes in their concentrations.17
Filamentous cells of coli
A filament has been known for more than a century and yet some facts about it remain to be discovered. Certain bacteria naturally grow as filaments for example actinomycetes,25 but some bacteria such as E. coli turn into filaments under stress conditions.26 There are several types of stress, including, DNA damage, inhibition of cell wall synthesis, and the effect of cell division by expressing certain thermosensitive mutations. Therefore, it is possible to apply these stress conditions in the laboratory by adding certain chemicals at specific concentrations to cause filament formation.27
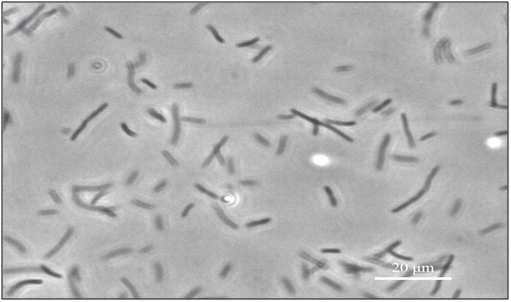
coli cells are normally rod shaped, as shown in Figure 3, but upon the addition of a b-lactam antibiotic, for example cephalexin,28 the cells replicate their DNA, as cell division is blocked. However, cell growth continues for several hours, before growth ceases and lysis occurs, resulting in long, multi-nucleoid and non-separated cell called filamentous E. coli (also known as snake), which is 50-200times longer than normal cells of E. coli, as shown in Figure 4.28–30 Although, they are longer, they still have the ability of movement,31 act as normal cells, and have the ability to synthesize flagella and express genes.18,32 The absence of a dividing wall (septum) of filaments allows tiny molecules to diffuse throughout the cell’s interior.33 Filaments are a research priority that have been used to study fields including chemotaxis, swimming, and cell division and growth.18,34–36
The experiment and its aim
As our lab is interested in the effects of cell velocity on its behaviour, this project will give baseline data on the biology of filamentous forms of E. coli cells that will later be used in further experiments. It is important to note that stress conditions induce filaments formation,27 and this change of form might affect bacterial chemotactic behaviour.
Our objective is to compare the swimming velocity in normal E. coli (wild type strain AW405) to filamentous cells of E. coli (treated with cephalexin antibiotic) in the absence of a Chemoattractant (control) and when chemotaxis conditions are applied (using L-Serine amino acid as an attractant). The experiment will be conducted twice by applying the same procedures to ensure validity and accuracy of results. The velocity was measured over time by taking videos every 30minutes for 4hours of both kinds of cells, using a Dunn chamber and a20X objective lens, under two different conditions:
Experimental hypotheses
Bacteria
The bacteria used for this chemotactic study was wild type E. coli strain AW405,37 provided by NCIMB Ltd.
Media
Minimal media preparation was conducted according to the Vogel-Bonner method.38 As for carbon and energy source, 60% (w/w) DL-lactate was used, and the following amino acids were present in the nutrient solution (L-histidine, L-leucine and L-threonine).
Growth conditions
Bacteria were stored at -70°C, and later placed in 25ml of minimal media in a 100ml flask and shaken at 200rpm on (InnovaÒ44) to grow overnight at 33°C. 1ml of these cells were inoculated in 25ml of minimal medium and shaken again at 200rpm on (InnovaÒ44) at 33°C in a 100ml flask for 2hours. To induce filament formation, normal cells are treated with 144mg of cephalexin in 24ml of cell culture flask, giving a final concentration of 6mg/ml.
Bacterial Concentration
Optical density of cells is measured at a wavelength of 600nm by the spectrophotometer (UNICAM 8625). Through trial and error, the standard optical densities of cells and filaments for clear microscopic observations have been found to be as the following: the first experimental group is the normal E. coli cells at 0.11-0.18, and the second experimental group is the filamentous E. Coli at 0.1-0.14.
Chemicals
L-serine (10-2M) was evaluated by Mesibov & Adler7 assay. It was used in all experiments as an attractant, and was provided by (Duchefa ). It was selected to prepare 30ml of 10-2M of L-Serine, which was decided to prepare 10-1M of L-Serine, and then dilute the solution to lower concentration, to avoid mechanical errors in weighing. Therefore, the preparation was done in the following steps:
Preparing 30ml of 10-1M L-Serine (molecular weight is 105.1g/mole), the concentration can be calculated as follows
Then, in the lab 0.3153g of chemical powder was added onto a flask containing 30ml distilled water, and shaken very well using vortex (IKAÒLab Dancer) until powder dissolved completely.
Preparing 30 ml of desired concentration 10-2M L-Serine, can be calculated from step 1 solution as follows
From the above equation, in the lab 3ml of 10-1M of L-Serine that was prepared in step 1, was added to a flask containing 27ml of distilled water, and shaken very well using vortex. Finally, the solution is stored in a lab refrigerator.
Dunn chamber
In 1991, Zicha et al.39 created the Dunn chamber for the purpose of chemotaxis long-term studies, especially for slow moving cultured like fibroblasts. The basic design idea was to improve the gradient stability as instabilities were caused by a large number of causes, such as mechanical loss, thermal effects, small alterations in the effective volumes of the wells, or a slight change to the height of the diffusion gap, which can all result in alterations of the gradient formed at the gap. The Dunn chamber is a device constructed at the Randall Institute to overcome the flaws of the Zigmond Chamber, which was the standard direct-viewing chemotaxis device.40 The Dunn chamber is built from a Z3 Helber bacteria counting chamber41 and is made entirely of glass that is 1 mm thick, which is optimal for microscopic vision as the diffusion gap’s height can be determined prior to the experiment using optical machining, and ensures a 20μm gap height is maintained when correct assembly of the chamber takes place as shown in Figure 5.39,42

The layout of the bridge and wells is concentric with the bridge being ring shaped and is 1mm wide,41 as opposed to the linear pattern in Zigmond’s,40 which decreases the possibility of flexure, providing a more secure placing of the cover slip and avoiding linear bridge end effects. The central well in the Dunn chamber is completely blind, as compared to other designs in which it is either wax closed or remains open such as the Zigmond chamber, which also ensures a constant volume. Forced overflow could affect the diffusion gradient and is prevented from passing over the bridge even in the flexed state provided the culture medium is air-bubble free, this is an advantage of the central well in this design. However, the inner well medium cannot be changed while the experiment is ongoing.39,42
In the lab, the preparation of the Dunn chamber was evaluated by Zicha et al.42 assay. However, 140ml of bacterial culture were taken from a 25ml culture and pipetted into the inner well of Dunn chamber, and a cover slip was used to completely seal the inner well. The outer well is sealed as well but a filling slit is left open. Light pressure was applied to the cover slip to remove excess contents of the inner well. An absorbent tissue paper is inserted through the filling slit to remove excess contents that have oozed into the outer well. 70mg of either the control (minimal media) or L-serine (attractant) were pipetted into the outer well, ensuring no bubble formation. Another cover slip is used to cover the filling slit and absorbent tissue is used to drain excess fluid. Petroleum jelly was then applied to the edges of the cover slip to seal it into the chamber.
The formation of chemical gradient
The study starts after steady state has been reached, when the chemical gradient formation is complete, which takes 30 minutes, calculated according to the Zicha et al.39 assay method, which states that due to the stable and precise dimensions of Dunn chamber, the formation of gradient could be accurately assumed if the diffusion coefficient was known for the substance. The diffusion is a mechanism of mass transport, which is thought to be in the bridge region only, whereas the contents of the two wells are kept stirred by convections currents. Theoretically, the diffusion is the same as the diffusion of a hollow circular cylinder and solutions for steady and non-steady states within the wall, which are evaluated by Crank.43 The concentration of steady state from the centre of the chamber as a function of distance, r, is:
Where,
= The concentration of the inner well.
= The concentration of the outer well.
a = The inner radii of the bridge.
b = The outer radii of the bridge.
Due to annularity of the bridge, the concentration gradient is not precisely linear, as a higher concentration in the outer well makes it a little convex, with very small deviation from linearity.
Microscopy
Chemotactic study was performed using Dunn chamber, attached to an inverted microscope (ZEISS, AXIO Vert.A1) using x20 magnification at 33°C thermal stage with phase contrast for bacterial motility videos and x10 magnification at 33°C thermal stage with phase contrast for bacterial gradient formation photos (Linkam T-95).
Camera and videos
In all experiments, the prepared slide was left for 30minutes for chemical gradient formation. The duration of chemotactic study was done over 4hours with a chosen standard time of 30minutes for observation and video recording of bacterial motility in all experiments. A camera was used (Pixelink PL-B686CF), with the specific settings (1500frame/video, 50frame/sec and 792x600pixels) chosen and being consistent in all videos and photos.
Data analysis
All video recordings of chemotactic study were manually tracked using Image J program (MTrackJ plugins) to quantify bacterial velocity. Then, the data given were analyzed using Statistical Package for the Social Sciences IBMÒSPSSÒStatistics (Version 2.0).
Normal cells of E. coli
Movement in normal cells: L-Serine amino acid was attractive to normal E. coli.37 After the addition of 1x10-2 M of L-Serine (an attractant) in the outer well, normal cells of E. coli (not treated with cephalexin) showed continuous running and blocked tumbling. However, a process called adaptation followed this process, in which the bacteria returned to tumbling and running.18 Whereas, in the absence of L-Serine and presence of minimal media in the outer well, normal cells of E. coli showed continuous running and tumbling. The total distance travelled by normal cells of E. coli might be different from the distance travelled by filaments in a set time period as shown in Figure 6.
Microscopic observations: As mentioned earlier, our goal was to compare the velocity between filaments and normal cells of E. coli under the effect of chemotactic stimuli. Therefore, to reach our objective, identical conditions and setting parameters were applied to all the videos and photos that were taken (see materials and methods). However, it is important to note that observations started in all experiments after chemical gradient formation (30 minutes) and the times mentioned later will exclude this period. The first video was taken in all experiments after chemical gradient formation. All observations of cells movements were seen using a 20X magnification lens and whole bridge view using 10X magnification lens.
Without an attractant: On the other hand, in the absence of L-Serine (Figure 7), 10 minutes after chemical gradient formation, it had been noticed that most of the normal cells were motile and concentrated near bacterial culture side. The movement of bacteria was slow, almost 20µm/s, and the concentration of cells was more near the bacterial culture side up to 1 hour. After 1.5 hours, the E. coli moved and was distributed everywhere on the bridge, but cells were more concentrated in the middle of the bridge in Dunn chamber. After 2.5 hours most of the normal E. coli cells were attaching themselves to surfaces (cover slip), therefore bacterial movement could barely be seen.

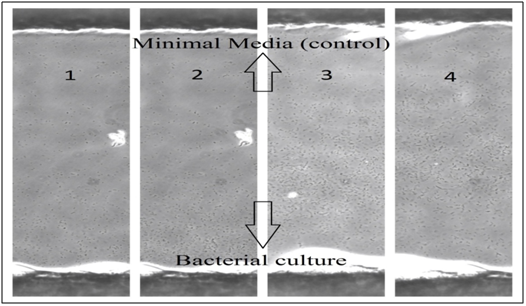
With an attractant: Figure 8 shows 4 different times of E. coli movements on the bridge in Dunn chamber, which illustrates the bacterial population presence (concentration) over time and how the bacteria moved in the presence of L-Serine (an attractant). Ten minutes after chemical gradient formation, most of the normal cells were motile and concentrated near bacterial culture side, and after 1 hour they started to form a bacterial gradient by moving toward L-Serine (attractant), which means the E. coli concentration will move to the other side because of the effect of chemotactic stimuli by L-Serine. This bacterial gradient of normal E. coli was clearly seen after almost 2.5 hours. It is clear to notice in this group (with attractant) that E. coli cells were more active and moved faster than the other group (without attractant). On the bridge, the bacteria on L-Serine side remained alive and appeared to be more active for a much longer time (up to 3 hours) than the bacteria on bacterial culture side (up to 2.5 hours), which have mostly stopped moving, and interestingly found to be attached to the cover slip, it was also noticed that they tend to attach to the glass surface much quicker in the absence of a Chemoattractant.

Filamentous E. coli
The growth and movement in filaments: Our results showed that after the addition of cephalexin to E. coli cells (see materials and methods), cell division stopped and cell growth continued and filamentous cells were produced.29 This process of growth development in minimal media could occur up to 6 hours after the addition of cephalexin. It is noticed that the cell division did not require flagellar synthesis, which was randomly distributed over the cell body. L-Serine amino acid was attractive to filamentous E. Coli. As shown in Figure 9, with L-Serine addition, the filaments showed the ability to be motile and ran continuously. Also here, the chemotactic response of filaments was followed by adaptation. The direction of running was either in the opposite or the same direction before stopping. Without the L-Serine addition, they were running and stopping instead of tumbling because of their elongated body. The motile filaments appeared to be running and stopping continuously with frequent stopping.18 As shown in Figure 6, the total travel distance of filaments is longer than that of normal cells of E. coli. The filaments were observed for 4 hours after the addition of cephalexin.

Microscopic observations: The microscopic settings used with filaments are exactly the same settings used with normal E. coli cells (see materials and methods). It had been noticed during the experiments that both normal and filamentous cells of E. coli have almost the same response to the chemotaxis condition with attractant18 and normal condition without attractant (control). Therefore, the observations were almost similar to each other in normal and filamentous cells. The movement of filaments over the bridge resembled the movement of normal cells in Figures 7 & 8.
Without an attractant: On the other hand, in the absence of L-Serine, it had been noticed that most of the cells were motile and present near the bacterial culture side at zero time, in which the velocity of filaments was approximately 28 µm/s. The filaments were distributed over the bridge but were more present near bacterial culture side in the first hour. After 2 hours, the filaments distributed more over the bridge and were present more in the middle of the bridge of Dunn chamber. After 3 hours, most of the filaments started to die, and attached to the cover slip, therefore filaments’ movement could barely be seen.
With an attractant: After chemical gradient formation, most of the filaments were motile and present (concentrated) near bacterial culture side, and after 1 hour they started to form a bacterial gradient by moving toward L-Serine (attractant), which means the filaments concentration will move to the other side because of the effect of chemotactic stimuli by L-Serine. This bacterial gradient of filamentous E. coli was clearly seen after almost 2 hours. On the bridge, the filaments on L-Serine side remained alive and appeared to be more active for a much longer time (up to 4 hours) than filaments on bacterial culture side (up to 3 hours), which have mostly died, and found some to be attached to the cover slip. Most filamentous E. coli cells start to become less motile after 3.5 hours in the bacterial side of the bridge.
Velocity versus time
Chart 1 illustrates the velocity of filaments and cells over 4 hours with and without an attractant. Upon the addition of L-Serine, normal cells of E. coli showed a fluctuated decreased in velocity over time (4 hours). Normal cells ran continuously toward the attractant, and then adapted to the environment, and the effect of chemotactic stimuli decreased gradually. But, in the absence of L-Serine, the velocity showed to be steady at zero time to 2 hours and then dropped. Normal cells ran and tumbled with frequent tumbling. Overall, cells with an attractant appeared to move faster than cells without an attractant.
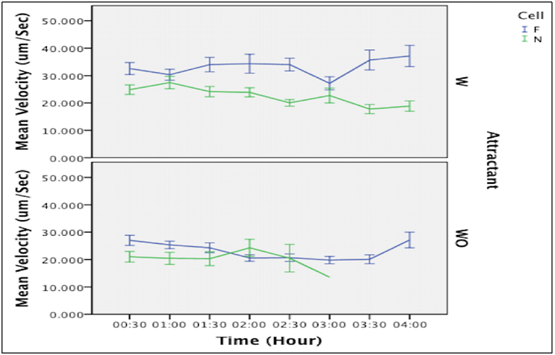
Nevertheless, after the addition of L-Serine, filaments showed a fluctuated increase in velocity. Filaments ran continuously and stopping was blocked, after that adaptation to chemotactic stimulation occurred. Whereas, without the addition of L-Serine, they showed a decrease in velocity with a slight increase at 3.5 hours. Filaments ran and stopped with frequent stopping. However, it is obvious that L-Serine addition increased the velocity of filaments.
In comparison between the velocity of both filaments and normal cells, our result data showed that filaments moved faster than cells at most of the time in both conditions with and without an attractant as shown in Chart 1. The mean velocity illustrated in Table 1 shows the difference between the filaments and normal cells in both conditions (with and without an attractant).
Experimental hypotheses
As mentioned above, the experiment was repeated twice, and permitted the same exact technique. Tables 2 & 3 show the number of bacteria tracked in each experiment, in which 20 was the maximum number of bacteria tracked per video. Experiments with bacterial numbers less than 20, demonstrate a lower number of moving bacteria during the video. This raw data was transferred to (SPSS) program to be analysed.
E. Coli |
L-Serine |
The Mean Velocity |
Cells |
With |
23.12 |
Without |
21.04 |
|
Filaments |
With |
33.14 |
Without |
23.12 |
Table 1 Illustrates the mean velocity (calculated from the sum of values recorded from the two experiments) of cells and filaments in both conditions with and without L-Serine (an attractant)
Time |
Normal E.coli (AW405) |
|||
|---|---|---|---|---|
Without attractant (1) |
Without attractant (2) |
With attractant (1) |
With attractant (2) |
|
30 Mins |
8 |
20 |
20 |
20 |
1 Hour |
4 |
20 |
20 |
20 |
1.5 Hours |
9 |
20 |
20 |
20 |
2 Hours |
1 |
13 |
20 |
20 |
2.5 Hours |
No motility |
4 |
20 |
20 |
3 Hours |
No motility |
1 |
1 |
20 |
3.5 Hours |
No motility |
No motility |
No motility |
20 |
4 Hours |
No motility |
No motility |
No motility |
20 |
Table 2 Shows the number of bacteria tracked in each experiment of normal cells of E. coli
Time |
Filamentous E.coli (AW405) |
|||
|---|---|---|---|---|
Without attractant (1) |
Without attractant (2) |
With attractant (1) |
With attractant (2) |
|
30 Mins |
20 |
20 |
20 |
20 |
1 Hour |
20 |
20 |
20 |
20 |
1.5 Hours |
20 |
20 |
20 |
20 |
2 Hours |
20 |
20 |
20 |
20 |
2.5 Hours |
20 |
20 |
20 |
20 |
3 Hours |
20 |
20 |
20 |
20 |
3.5 Hours |
20 |
20 |
20 |
20 |
4 Hours |
20 |
20 |
20 |
20 |
Table 3 Shows the number of bacteria tracked in each experiment of filamentous cells of E. Coli
The first null hypothesis: Type-III two-away ANOVA was conducted to compare the velocity between filamentous and normal E. coli cells in the absence of an attractant, which indicated a significant difference between them (F(2,1006)=105.01, MSE=69.864 P<0.001). The result indicates a significant difference between filament and normal E. coli cells, which is that the filament cells have a faster movement (the velocity mean is 23.12mm/s) than normal cells (the velocity mean is 21.04mm/s) (Chart 2). Therefore, the first null hypothesis was rejected. Where the velocity mean of filament cells is 23.12mm/s and the velocity mean of normal cells is 21.04mm/s (Chart 2).
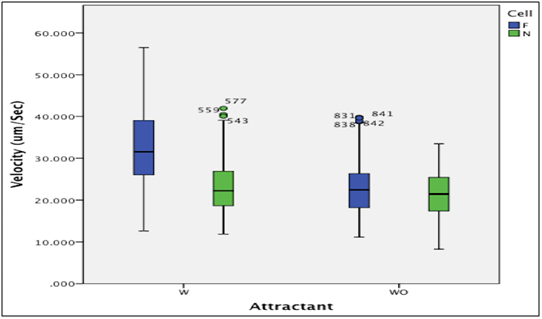
The second null hypothesis: Type-III two-away ANOVA was conducted to compare the velocity between filaments and normal cells of E. coli in the presence of an attractant (L-Serine), which indicated a significant difference between them (F(2,1006)= 127.04, MSE= 68.511 P<0.001). Therefore, the second null hypothesis was rejected. It is obvious that the chemotactic stimulation effects on filament cells was much more than on normal cells. Where the mean velocity of filament cells is 33.14mm/s and the mean velocity of normal cells is 23.12mm/s (Chart 2).
To study the velocity of filaments and normal cells of E. coli under chemotactic conditions, cells were placed in the inner well of Dunn chamber and 1x10-2M of L-Serine was added to the outer well. Cephalexin, a b-lactam antibiotic was added to normal E. coli cells to produce filaments. The bridge between inner and outer wells was used to measure the velocity of immigrant cells and filaments toward the attractant. The normal cells ran continuously and as the influence of Chemoattractant decreased, it resulted in adaptation mechanisms and cells returned to running and tumbling. Filaments on the other hand, were motile (Figure 8), and also ran continuously followed by adaptation mechanisms resulting in retuning to running and stopping instead of tumbling because of their elongated body. The normal cells and filaments have the same chemotaxis responses. The times required for normal cells to adapt were much longer than filaments in the presence of Chemoattractant (240s compared to 90 s for the filaments).18
Therefore, in the presence of L-Serine in the outer well, prolonged running in a biased favourable direction toward (L-Serine) was induced (Figure 2), which explains the high velocity of cells and filaments. Whereas, in the presence of minimal media in the outer well (control), cells moved normally (running and tumbling alternately); tumbling while moving of E. coli caused a slow motion, which explains the decreased velocity in the absence of the attractant. Likewise, the stopping of filaments had the same influence on their velocity. Figure 6 is inspired from our lab observations, which illustrates the difference in total distance travelled by normal cells and filaments and their actual displacements on the bridge of Dunn chamber. The total distance travelled by normal cells of E. coli might be different from the total distance travelled by filaments in a set time period. Although, in a set period of time, normal cells might travel a longer total distance, the displacement is shorter. On the other hand, filaments might travel a shorter distance, but the displacement is longer in the same period of time set for normal cells. The total distance travelled in normal cells of E. coli results from frequent change in their direction after each tumble, whereas filaments turn less frequently as they tend to continuously run in the same direction. From these observations, it could be said that filaments travel faster from start to end points in a set period of time compared to normal cells that travel slower from start to end points in the same set period of time.
To explain the increased velocity of filaments shown in Chart 1, which should have been decreased due to adaptation mechanism, our suggestion is they stop less frequently and tend to travel in a straighter path as explained above; also the huge elongated body (up to 50times bigger than normal cells ≈100mm long) moves a longer distance per run. On the other hand, the normal cells of E. coli tend to tumble more than filaments stop, use an indirect path, and travel a shorter distance per run compared to filaments. As mentioned earlier, filaments division is blocked upon the addition of cephalexin and body size increases gradually over time, which means the distance travelled per run is increased as well, so, this explains the increase of the velocity in filaments over time. Importantly, the two conditions with and without L-Serine showed the same increase over time, which supports our suggestion. Further study of size and velocity is needed, as it will lead to more accurate results.
In comparing the velocity of filaments and normal cells with L-Serine, our data findings show that the filaments are faster (33mm/s) than normal cells (23mm/s). Our findings do not match those mentioned in Maki et al.18 study. Their data show the opposite result, in which the normal cells (19mm/s) were faster than filaments (11mm/s). This leads us to more than one probability (i) a mechanical or calculation mistake occurred during the conduction of experiments, (ii) the concentrations used in our study and their study were different, which might have affected the results, (iii) the methodology used in their study was not clear, and a spatial gradient was not created as was done in our study. The study of chemotaxis needs precise knowledge of chemical gradient formation, as cells need to sense the increase in concentration of the Chemoattractant to respond to its stimulation,44 this explains why Maki et al.18 results might be inaccurate, (iv) our technique using the Dunn chamber is used for the first time to conduct a chemotactic study on free swimming organisms and may need further tests to confirm its sensitivity. However, the normal cells in our study have a velocity of (23mm/s) which is almost the same velocity found in their study (19mm/s). This means that the determining the velocity of filaments using the Dunn chamber technique needs further studies.
The behavior of genetically modified nonmotile filamentous E. coli cells (carrying flaE hag mutations) bearing polyhooks to the flagellar motor instead of flagella was studied. To follow the flagellar motor’s direction of rotation, normal polyhook cells were fixed with glutaraldehyde to act as markers that were attached with anti-hook antibodies to the polyhooks, and examined by phase contrast microscopy. It was observed that polyhooks displayed altered spin bias in the presence of Chemoattractant (L-Aspartate) suggesting attractant inactivation and a one-third decrease in chemotactic signal strength every 2μm as it diffuses throughout the cytoplasm.34 The chemotactic signal diffused through the cytoplasm goes through dephosphorylation and is mediated by Che Y-P.45 It is unlikely, that Che Y-P has the ability to reach flagella from the cell poles all along the cell body in filamentous cells.18
Through florescence microscopy, it was found by Maki et al.18 that filaments contained MCPs in their poles and along the cell body. The amount of MCP clusters found along the cell body however was greater than that found at the filament poles. As CheW deletion retracts MCP localization, it may be concluded that it is necessary for both polar and nonpolar MCP cluster formation.
In normal E. coli cells, MCPs are mainly found at the poles of the cells and chemotaxis depends on CheY diffusion from these MCPs to the randomly placed flagella.46 Time of initiation of response in normal sized cells is about (0.1s) which can be attributed to CheY diffusion, as its diffusion constant is sufficient to account for the response.47,48 If it were assumed that MCPs of filaments are only present at the poles, increased cell length as well as short CheY-P half-life would impose difficulties in conducting proper chemotactic reposes.34,49 However, our results showed that E. coli filaments have similar chemotactic responses to those of normal size, which corresponds with work by Maki et al.18 Some of the anticipated strains on the filamentous cells due to their size arise from the presence of concentrated lateral MCP clusters along the body of the cell. It is hypothesized that Chemoattractant and chemorepellents are sensed by filamentous E. coli through MCP clusters at the poles and along the cell body, and normal chemotactic responses result from this mechanism.
Since CheY-P diffuses from an MCP cluster to a close by flagella, a stimulus of uniform concentration is thought to induce a response in all flagella along the filament, which are within a close proximity to the MCP cluster (within 1-2mm). If the stimulus concentration between filament ends differs notably then it is a non-uniform stimulus and the response is predicted only at the nearby flagella. Segall et al.34 observation supports this theory as it states that altered bias is displayed by polyhooks in the local vicinity of applied attractant.
As a result of complex adaptation mechanisms, cells that come in contact with a repellent immediately have the chemoreceptors send a signal for the kinase to be activated and a responsive motor response is initiated, a few seconds later however, they resume their swimming in the same prestimulus behavior even with the repellent’s presence in the surrounding environment. Alterations in methyl esterification of the glutamate side chains of receptor proteins are involved in the adaptation techniques.50
Each of the receptors has a conserved signalling domain, at which specific residues of glutamate undergo methylation by CheR methyltransferase, and undergo demethylation by CheB methyltransferase. The regulatory domain of the demethylation process is homologous to the CheY response regulator, therefore it is subject to CheA kinase phosphorylation and activation. Demethylation however, induces a receptor state that deactivates the kinase, resulting in the demethylation process acting as a negative feedback mechanism used to develop the adaptation of bacteria to its repellents. Likewise, adaptation to attractants is developed by an increase in methylation of the receptor, and the activation of kinase. Methylation is a mechanism that ensures a constant swimming behavior for bacteria under varying environmental changes.51 So far, glutamate reversible methylation seems to be distinctive to bacterial chemoreceptors only, therefore this receptor family has been named methyl-accepting chemotaxis proteins (MCPs).24
Dunn chamber is designed to study chemotaxis, and was used before on leucocytes39,41,42 and cancer cells.52 It is used for the first time to measure chemotaxis ability on free-swimming organisms. The experiment duration is short for our study, it has been suggested for more studies with longer times. As for troubleshooting, it had been noticed that the bacteria die during the experiment, which cause a delay for a few days. It could be due to either an error that occurred during the bacterial culture or the technique applied to prepare the Dunn chamber. However, this problem needs a longer time of investigation to find the cause and its solution.
The Dunn chamber method has many advantages in chemotaxis study including: Easy handling, easy detection of cell movement and the advantages of Dunn chamber construction (see materials and methods). The disadvantages observed during the experiment include:
The addition of cephalexin b-lactam antibiotic to normal cells induced filament formation. Upon this addition, cells replicated their DNA, as cell division is blocked but cell growth continued for several hours. Compared to the small size of normal E. coli cells, filaments were huge and elongated (up to 50times longer). Despite being longer, they still had the ability to move, synthesize flagella and express genes. Dunn chamber is a good technique that was used in the study of chemotaxis of E. coli cells. In the absence of a Chemoattractant, the normal cells ran and tumbled alternately, whereas, the filaments ran and stopped alternately. In the presence of a Chemoattractant, both kinds of cells ran continuously, followed by adaptation mechanism returning to un-stimulated condition. The normal cells and filaments showed the same ability of motility and chemotaxis. Our findings indicate that the filaments were faster than the normal cells. The total distance travelled by normal cells of E. coli is shorter than the distance travelled by filaments in a set time period. Microscopic observations of cells and filaments showed similar movements. Our suggested explanation to the increased velocity of filaments over time is that they stop less frequently and tend to travel in a straighter path; also their elongated body might increase the distance per run. This increase in cell size is directly proportional to the increase in the each distance travelled per run. Further study of size and velocity is needed, as it will lead to more accurate results.
First and foremost, I would like to thank my supervisor Dr Stuart Humphries, for his support, valuable guidance, and advice throughout this project. He inspired me greatly to work in this study and his willingness to motivate me contributed tremendously to my project. I would also like to thank dròscar Guadayol i Roig for his help and assistance in lab work. I would like to thank everyone in the physical ecology lab for creating a lovely environment to work in. Furthermore, I would like to thank University of Hull for providing us with a good environment and necessary facilities to complete this project. Lastly, an honourable mention goes to my charming wife, Wala Turkistani, my lovely family and friends for their understanding and support. Without the help of the particular mentioned above, I would have faced many difficulties while doing this project.
The author declares no conflict of interest.

©2015 Niaz. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.