MOJ
eISSN: 2374-6912


Research Article Volume 2 Issue 2
1Department of Biology, University of South Alabama, USA
2Department of Pharmacology, University of South Alabama, USA
Correspondence: Glen M Borchert, Department of Biology & Pharmacology, Assistant Professor of Biology & Pharmacology, University of South Alabama, Alabama, USA, Tel (251) 4607310, Fax (251) 4148220
Received: April 27, 2015 | Published: May 22, 2015
Citation: Cardin SE, Perry TJ, Polska CJ, et al. MicroRNA sponge production using PCR-based concatemerization of short DNA oligonucleotides. MOJ Cell Sci Rep. 2015;2(2):50-53. DOI: 10.15406/mojcsr.2015.02.00025
Micro RNAs (mi Rs) are small, non coding RNAs encoded within our genome that regulate gene expression by silencing messenger RNA (mRNA) transcripts. Since being discovered in humans in 2001, much has been learned concerning the many cellular activities that mi Rs can affect. Mi Rs have been implicated in almost every cellular activity, including cell fate determination, stress response, metabolism, apoptosis, and carcinogenesis. Importantly, alterations in mi R activity due to mutation or mis expression have been repeatedly shown to result in tumorigenesis and disease. However, because of their relatively recent discovery, therapeutic tools to suppress mi Rs continue to be developed. One extremely promising way to inhibit mi Rs is to use mi R sponges which consist of ~15 mi R “target” sites that can specifically bind and inactivate particular mi Rs. Unfortunately, mi R sponge production has proven to be problematic thus far as current production methods involve either costly commercial synthesis or low throughput ligation-based cloning. However, we have recently developed a novel, PCR-based method that allows sponge production in a rapid, high throughput manner. In all, PCR amplification, cloning, sequence confirmation and large scale production are readily achievable in less than two weeks and at a significantly lower cost than current methods.
Keywords: micro rna, mi rna, mi r, mi r inhibitors, mi r sponge, pcr
Bp, base pair; CE RNA, competing endogenous rnas; Circ RNA, circular rna; Ci RS, circular rna sponges; LINC RNA, long intervening non coding rnas; LNA, locked nucleic acid; Mir, micro rna; MRNA, messenger rna; NT, nucleotide; PCR, polymerase chain reaction; Pre-Mi R, precursor micro rna; Pri-Mir, primary micro rna; RISC, rna induced silencing complex; SIRNA, small interfering rna; UTR, un translated region
Micro RNAs (mi Rs) are small, ~20 nucleotide (n t) non-coding RNAs that act as regulators of gene networks. Since being discovered in Caenorhabditis elegans in 1993, studies have identified over 2,500 unique human mi Rs and over 35,000 mature mi R products across species. Mi Rs are widespread in higher eukaryotes and similar in function to small interfering RNAs (si RNAs), in that they silence specific messenger RNAs (mRNAs). Generation of mi R from endogenous hairpin-shaped transcripts occurs in two main stages: within the nucleus and without.1 First, the mi R gene is transcribed within the nucleus by either RNA Polymerase II or III producing a long primary molecules, called a pri-mi R typically composed of several thousand nts.2 The pri-mi R is then cut by Drosha, yielding a ~70nt long precursor stem loops (pre-mi R). Next, the pre-miR hairpin is exported to the cytoplasm by exportin-5, where Dicer cuts it down to a ~22nt duplex. One strand is degraded and the other is retained as the mature mi R that associates with the RNA induced silencing complex (RISC).1,3 From there, the complementarity between the 6-8nt ‘seed’ region of the mi R and the ‘seed match’ region on the mRNA determines the fate of the mRNA.4 Importantly, mi R binding does not have to be perfect in order to have an effect. When a mi R seed binds to the mRNA seed match, it can result in one of two things: mRNA cleavage in the event of perfect full-length mi R complementarity, or translational repression if the match is imperfect. Importantly, a single mi R can influence the activity of numerous mRNAs, an ability which in turn allows mi Rs to form intricate regulatory networks.5
Altered activities of several mi Rs due to mutation or mis expression have now been shown to result in disease onset and/or tumorigenesis.6-9 While miR mis regulation is implicated in numerous malignancies, therapeutic tools for mi R suppression have only recently been developed.6 Commercially available mi R inhibitors fall into three major categories: antagomirs,10 synthesized sponges,11 or antisense inhibitors.12 Antagomirs, better known as LNAs (locked nucleic acids) or anti-mi Rs, are chemically modified nucleic acids that strongly bind a target mi R by traditional complementary base pairing. The thermodynamic stability of LNA binding keeps them tightly associated with their targets, but also allows for imperfect base pairing at high temperatures. Unfortunately, LNAs carry the risk of off-target effects13 making them less desirable candidates for therapeutic endeavors despite their demonstrated utility as a tool for basic research.11 In contrast, antisense inhibitors (such as Ambion’s mirVana) contain an unmodified RNA strand (which binds targets with normal affinity) and a chemically modified strand that cannot associate with RISC. Although this class of inhibitors is not hindered by the potential toxicity associated with LNAs, these antisense inhibitors can still prove problematic for many researchers in terms of the cost associated with their production.12 Significantly different in design than either of the synthetic inhibitors, the third type of mi R inhibitor currently employed is the mi R sponge. Mi R sponges are RNA transcripts with repeated binding sites forming a concatemer of targets for a specific mi R (Figure 1). Sponges can be constructed with repeats of any sequence, but sponges with central mismatches, called a central bulge, have been shown to be more effective at inhibiting mi R function than perfectly complementary sponges.11 To date, sponges have been synthesized by either commercial synthesis or T4 ligation reactions. While more cost effective than commercial synthesis, due to the linear nature of their reaction mechanism, T4 ligation reactions are notoriously low throughput and typically require large amounts of starting material to produce relatively low product yields.14
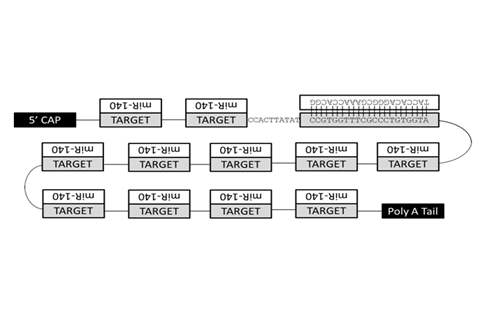
Excitingly, we have developed an entirely novel, rapid, PCR-based methodology that greatly facilitates sponge production. The concatemerizing PCR technique we present here represents a cost-effective and practical method for mi R sponge production. Instead of time-consuming ligation reactions that produce little overall yield, concatemerizing PCR utilizes amplification steps and exponential growth to quickly generate large quantities of sponge amplicons that consist of an easily user-defined number of mi R binding sites. Importantly, as prominent roles for mi Rs in disease development and progression continue to be described7-9,15 knocking down mi Rs in the laboratory and clinic has become an increasingly significant endeavor, and the method of sponge production described here constitutes an innovative, inexpensive, high yield means of mi R inhibition.
Primer design
Approximately 40 nucleotide forward primers were constructed with GGTGAATATA at both the 5’ and 3’ ends flanking an internal sequence to be repeated and concatemerized. Reverse primers were ordered exactly as the reverse complement of the forward primer allowing concatemerization. Each sponge was concatemerized with the following primers:
miR140 sponge:
GGTGAATATATACCACAGGGCGAAACCACGGGGTGAATATA,
TATATTCACCCCGTGGTTTCGCCCTGTGGTATATATTCACC
SPG1:
GGTGAATATATTAATACGTGACCTATTAATGGTGAATATA,
TATATTCACCATTAATAGGTCACGTATTAATATATTCACC
SPG2:
GGTGAATATATAGCTTATCAGACTGATGTTGAGGTGAATATA,
TATATTCACCTCAACATCAGTCTGATAAGCTATATATTCACC
SPG3:
GGTGAATATACTAATACTGCCTGGTAATGATGAGGTGAATATA,
TATATTCACCTCATCATTACCAGGCAGTATTAGTATATTCACC
SPG4:
GGTGAATATACTAATACTGAAAGGTAATGATGAGGTGAATATA,
TATATTCACCTCATCATTACCTTTCAGTATTAGTATATTCACC
SPG5:
GGTGAATATATGGGAGGGGAGAGGCAGCAAGCAGGTGAATATA,
TATATTCACCTGCTTGCTGCCTCTCCCCTCCCATATATTCACC
SPG6:
GGTGAATATATGGGAGGGGCACGGCAGCAAGCAGGTGAATATA,
TATATTCACCTGCTTGCTGCCGTGCCCCTCCCATATATTCACC
SPG7:
GGTGAATATACATGCTGACCTCCCTCCTGCCCCAGGGTGAATATA,
TATATTCACCCTGGGGCAGGAGGGAGGTCAGCATGTATATTCACC
SPG8:
GGTGAATATACATGCTGACCAAACTCCTGCCCCAGGGTGAATATA,
TATATTCACCCTGGGGCAGGAGTTTGGTCAGCATGTATATTCACC
Concatemerizing PCR
PCR amplifications were performed in 50μL reactions at standard concentrations [10μL of Bioline 5xMyTaq buffer (comprised of 5.0mM dNTPs, 15mM MgCl2), 1.0μL Taq (Bioline, USA, Inc,. Randolph, MA), 0.2μm of each primer].The following cycling parameters: 94°C–2:00min, (94°C-30s 45°C-10s, 72°C-30 s)×2cycles, (94°C-30s, 40°C –30s, 72°C–30s)×29cycles, 72°C-1:00minute, 4°C -or ever, were utilized. Following this, resulting amplicons were separated on a 1% agarose gel and desired sponge length was extracted. See “Supporting Information - Supplemental Protocol” for additional details.
Vector cloning
The band corresponding to desired sponge length was excised from the appropriate lane at ~400 bp. Gel extractions were cloned into a pcr2.1-TOPOTA vector (Invitrogen). Concatemers were next excised from pcr2.1-TOPOTA and cloned into a mammalian expression vector (PCI, Promega, Madison, WI) using EcoR1 restriction sites which placed the sponge into the 3’UTR of the T7 promoter. Sponge orientation in PCI vector was determined by commercial sequencing (Functional Biosciences, Madison, WI). Sequencing primers for final constructs were: PCIF, TAATACGACTCACTATAGG; PCIR, AATAGCATCACAAATTTCAC.
To achieve sponge production, ~40 n t forward primers consisting of a simple linker (GGTGAATATA) flanking the 5’ and 3’ ends of an internal ~20nt mi R sequence to be repeated across the sponge were designed and synthesized commercially. Reverse primers (the exact reverse complement of the forward primer) were also ordered, thereby allowing concatemerization of the sequence complementary to the mi R by the mechanism depicted in (Figure 2A). PCR amplifications were initially performed in 50μL reactions at standard concentrations. Following amplification, PCR products were separated by gel electrophoresis, and regions corresponding to ~10 repeats (~400bp) were manually excised (Figure 2B) then cloned into the PCR 2.1-TOPOTA (Invitrogen) vector (Figures 2C & 3A). Next, to allow eukaryotic expression, sponges were removed from TOPO by the restriction digest of Eco RI sites immediately flanking both sides of the TOPO insertion, after which they were non-directionally sub cloned into a mammalian expression vector (PCI, Promega) linearized with ECORI (Figure 3B). Multiple colonies were isolated from resultant transformations and commercially sequenced (Functional Biosciences, Madison, WI). While expression vector sub cloning could alternatively be performed in a unidirectional manner following determination of orientation in PCR 2.1-TOPOTA by sequencing, this strategy allowed the simultaneous generation of both an expression vector producing a desired sponge and an identical expression vector producing the reverse complement of the sponge-an ideal transfection control. Importantly, Figure 2 demonstrates how a single round of PCR can be successfully employed to concatemerize single oligo nucleotide complements into markedly larger molecules tens of thousands of base pairs in length, from which any desired number of repeated miR target sites can be selected and utilized. Finally, we tested the versatility of our methodology by designing and attempting to amplify eight additional, unique sponges. Markedly, we found all eight primer sets robustly concatemerized under the conditions outlined here (Figure 4).
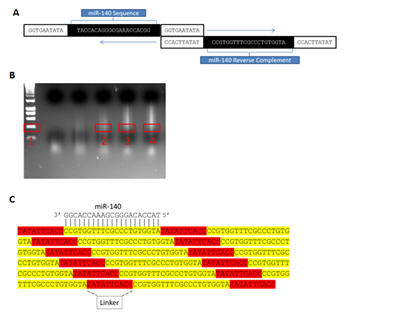
Figure 2 MicroRNA sponge production by concatemerizing PCR:
In summary, as mi Rs clearly play prominent roles in disease, development and normal cellular metabolism,15 knocking down mi Rs has become an important area of research. As such, the discovery of several types of naturally occurring mi R sponges strongly supports the application of engineered mi R sponges in mi R inhibition. Naturally expressed circ RNA or circular RNA sponges (ciRS) can contain up to 70 mi R mismatched target sites capable of binding multiple mi R families16 much like newly discovered (ce RNAs) which also demonstrate endogenous mi R sponge-like activity.17 Ce RNAs allow non-coding RNAs, such as mi Rs, to communicate via competition for mRNAs, with ce RNA function depending on mi R concentration. Ce RNAs contain binding sites for various mi Rs allowing them to regulate multiple mi R families.18 In addition, several other classes of non-coding RNAs may also function as endogenous mi R sponges (e.g. 3’ UTR RNAs and linc RNAs).19
Currently, sponges are synthesized by either commercial synthesis or T4 ligation reactions. While more cost effective than commercial synthesis, due to the linear nature of their reaction mechanism, T4 ligation reactions are notoriously low throughput and typically require large amounts of starting material to produce relatively low product yields.14 In contrast, the method of sponge production we describe here represents a highly efficient, enzymatic reaction utilizing Taq polymerase to readily generate potent mi R inhibitors that mimic endogenous sponges in a shorter time frame and at a substantially lower cost than existing production methods. Importantly, sponges can be constructed with repeats of any short sequence and designed to be perfect matches to mi Rs or instead to contain mismatches, or a central bulge (Figure 5), which has been suggested to be more effective at inhibiting miR function than perfectly complementary sponges.11 In addition, while we developed this PCR methodology to concatemerize miR target sites, we suggest this strategy could also be successfully utilized for other research purposes such as the concatemerization of transcription factor recruitment sites in expression constructs.
This work was facilitated by the University of South Alabama Department of Arts & Sciences, the South Alabama University Committee on Undergraduate Research (UCUR) and the Genetics Department at the University of Iowa. Funding for this work was provided in part by NSF CAREER grant 1350064 (GB) awarded by Division of Molecular and Cellular Biosciences, with co-funding provided by the NSF EPSCoR program and the Abraham A. Mitchell Cancer Research Fund (GB).
The author declares no conflict of interest.

©2015 Cardin, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.