MOJ
eISSN: 2374-6912


Research Article Volume 2 Issue 2
Department of Health and Human Services, American International Biotechnology, USA
Correspondence: Thomas Reynolds, Department of Health and Human Services, American International Biotechnology, 601 Biotech Drive, Richmond, VA 23235, USA, Tel 8043057648
Received: February 27, 2015 | Published: March 24, 2015
Citation: Budd WT, Harwich M, Bostwick GD, et al. Metagenomics analysis using next generation sequencing of vaginal samples from community practices in the us. MOJ Cell Sci Rep. 2015;2(2):16-25. DOI: 10.15406/mojcsr.2015.02.00020
Molecular-based Phylogenetic analyses, including the newly-completed Human Microbiomes Project and other next-generation sequencing (NGS) studies have revealed the complexity of the vaginal micro biome in health and disease. The healthy vagina tends to be dominated by hydrogen peroxide and lactic acid producing organisms, the most important of which are Lactobacillus species. The loss of these organisms and the rise of non-resident microorganisms can lead to a diseased state such as bacterial vaginosis (BV or vaginal bacteriosis). The new consensus generated by these studies is that BV is not a single disorder or caused by one etiological agent but rather it is a spectrum of symptoms clinically characterized by vaginal discharge, increased pH and malodor caused by an imbalance in the normal vaginal bacterial flora. NGS techniques allow for the capture of the entire vaginal micro biome in a single assay and create the ability not only to obtain a better understanding of the causes of BV but also lead to a viable clinical method for detecting it. In this study, we use next generation sequencing of 16S rRNA genes to obtain the vaginal micro biome from 270 vaginal swabs. Our analysis includes an in-depth characterization of bacterial communities identified, including quantitation and composition of the communities present in normal and BV samples. Our results show vaginal samples can be broken down into three distinct groups that segregate largely based on degree and type of Lactobacillus content and the presence or absence of G. vaginalis.
Keywords: metagenomics, bacterial vaginosis, next generation sequencing, gardnerella vaginalis, 16s sequencing
ATCC, american type culture collection; BHI, brain heart infusion; BV, bacterial vaginosis; CLIA, clinical laboratory improvements amendment; DNA, deoxyribonucleic acid; H2O2, hydrogen peroxide; LB, lutria broth; NaCl, sodium chloride; NGS, next generation sequencing; OD, optical density; PERL, pattern extraction and reporting language; PGM, personal genome machine; PID, pelvic inflammatory disease; rRNA, ribosomal ribonucleic acid; Spp, species; TSA, tryptic soy agar
Recent estimates from the human micro biome project estimate that the ratio of bacteria to human cells is 10:1.1 Evidence continues to grow suggesting that the composition and function of the micro biome is important for normal human development and continued homeostasis. Microorganisms tend to colonize at various sites and form complex communities adapted to their specific environment.2 The vagina tends to be dominated by hydrogen peroxide and lactic acid producing organisms, the most important of which are Lactobacillus species.3 The loss of these organisms and the rise of non-resident microorganisms can lead to a diseased state such as bacterial vaginosis (BV, or vaginal bacteriosis). BV occurs when healthy lactobacilli are replaced with a diverse polymicrobial population comprised of multiple taxa, including commensal facultative anaerobes and other pathogenic bacteria.4,5 Clinically BV is characterized by a spectrum of symptoms including vaginal discharge, increased pH, and malodor.
BV is the most commonly diagnosed vaginal infection in women of child-bearing age, accounting for 27-41% prevalence in Americans.6,7 It is common in postmenopausal women, with a prevalence of 23% that increases with age.8 BV is an important medical condition associated with more serious adverse outcomes such as acquisition of sexually transmitted infections/ HIV, pre-term child birth, and development of pelvic inflammatory disease (PID).9 Activities associated with an increased risk of BV include an increased number of male sex partners (>3 in 12months), having sex with a female in the previous 12months, previous pregnancies and cigarette smoking.10 Protected sex using a condom is considered a protective behavior and has been shown to limit the incidence of BV.
One of the greatest clinical challenges with BV is dealing with an extremely high rate of treatment failure (41% to 67% within months) despite use of oral or intravaginal antibiotics. Treatment failure is frustrating to patients and clinicians.11–13 There exist conflicting theories regarding the exact etiology of treatment failure. Some clinicians theorize that BV recurrence is a relapse of the same infection, while others believe it is not relapse but rather each occurrence is a new infection. Contributory risk factors for treatment failure include unprotected sex, sex with partners colonized by BV-associated bacteria, repeated bouts of BV, and use of an intrauterine device. As the frequency of BV increases with the number of sexual partners and unprotected sex, it was hypothesized that BV may be a sexually transmitted infection supporting the concept of re-infection.14 However, treatment of an infected woman’s partner did not statistically reduce the incidence of BV recurrence. Evidence suggests that BV recurrence is a relapse of the primary infection because there is a failure of vaginal re-colonization with healthy Lactobacillus spp. and continued colonization by anaerobic bacteria. Some of the BV-associated anaerobes, like Gardnerella vaginalis grow as a tenacious biofilm that adheres to the vaginal epithelium, making successful eradication more difficult.11 Several of the anaerobic bacteria associated with BV, such as G. vaginalis and M. curtisii, have low in vitro susceptibility to metronidazole, the drug most commonly prescribed to treat BV contributing to relapse if not all of the anaerobes are eradicated.15
The etiologic enigma has caused physicians and clinicians to diagnose BV based upon clinical criteria rather than on cultivation based tests and other standard microbial tests used when one organism is suspect. One such method for diagnosis of BV is through the use of Amsel’s criteria, where at least three of the following four conditions must be met: presence of a thin, homogenous discharge; vaginal fluid pH greater than 4.5; positive whiff test (generation of fishy odor when 10% potassium hydroxide is added to vaginal fluid); and microscopic presence of clue cells (sloughed epithelial cells coated with adherent bacteria).16 Amsel’s criteria cannot identify the etiologic agents of infection. A second clinical method for diagnosing BV is through use of the Nugent’s score which attempts to distinguish normal vaginal flora (lactobacilli are typically gram-positive rods) from BV-associated organisms (gram-negative organisms) by performing Gram’s stain on vaginal fluid.17 Neither of these methods is perfect or absolute; both tend to be highly subjective and depend on the skill and experience of the clinician processing the sample.18 Additionally, use of the Nugent score is based on an assumption of what the normal flora should be, which might not be totally accurate or complete.
Molecular-based phylogenetic analyses using 16S ribosomal RNA (rRNA) gene sequencing, including the newly-completed Human Microbiome Project and other next-generation sequencing (NGS) studies have revealed the complexity of the vaginal microbiome in health and disease.1,2,19 The 16s rRNA gene is an essential component of prokaryotic translation machinery. It is highly conserved and is a useful genetic target for microbiome type studies. The 16S rRNA molecule features areas of sequence conservation that can be used for primer design allowing for amplification of this target from various genetic backgrounds, yet there is also enough sequence diversity present to allow for the ability to identify organisms down to the genera and species level.20 Semiconductor sequencing technology (Ion Torrent) reduces the cost of per base sequencing and has been shown to be amenable to clinically relevant samples.21 Using a semi-conductor sequencer, for the first time we can deliver clinically actionable results in time for a physician to make treatment decisions.
In addition to identification of resident bacterial species, 16S ribosomal RNA gene sequencing using next generation sequencing (NGS) technology allows numerous improvements compared with existing methods of detection. Currently, clinical assays are capable of qualitative identification of bacterial species present in a complex community. It is known that many women are colonized with G. vaginalis but do not suffer symptoms of BV.18,19 Quantification of microbial community constituency will aid clinicians to more accurately assess the total microbiome of their patient, identifying those that are in need of treatment. Another benefit of an NGS based assay, is other gene targets can easily be easily added to an NGS panel. Virulence factors such as sialidase could enhance bacterial survival, invasion and/or tissue destruction; resistance genes can confer a selective advantage to bacteria making them immune to common treatments. The addition of these virulence factors and resistance genes can aid in diagnosis and treatment planning.22,23 In this study, we evaluated the composition of the vaginal microbiome by in-depth next generation sequencing of 270 women from community GYN practices in the United States. Our goal for this study is to better define the microbial communities inherent to normal and BV states, identify causative/ complementary bacterial communities, and show that vaginal microbiome analysis is a viable approach to the detection of BV in clinical samples.
Ethics statement
The data used in this study came from samples submitted to a CLIA approved clinical lab and was not collected specifically for this study. Researchers involved in this study were not aware of the participant’s identity, health status, or other personal information.
Media preparation
Brain Heart Infusion (BHI) broth (BD, Franklin Lakes, NJ), de Man Rogosa and Sharpe (MRS) broth (BD, Franklin Lakes, NJ), and Luria Bertani (LB) broth made according to the manufacturer’s recommendations unless otherwise noted. Corresponding solid media were also prepared by the addition of agar to 7.5%. Nutrient broth (BD, Franklin Lakes, NJ) and agar were prepared supplemented with 3% NaCl. Tryptic Soy Agar (TSA) + 5% sheep’s blood (BD, Franklin Lakes, NJ), CDC anaerobe blood agar (BD, Franklin Lakes, NJ), Human bi-layer tween (Remel, Lenexa, KS), Brucella blood agar (Hardy Diagnostics, Santa Maria, CA) and Cooked Meat Media (Hardy Diagnostics, Santa Maria, CA) were purchased ready to use from the indicated companies.
Bacterial growth conditions
Bacterial and fungal species (Table 1) were ordered from the American Type Culture Collection (ATCC) and grown in the indicated solid and liquid media. Cultivation of some anaerobes were routinely performed in BHI broth supplemented with 0.1% glucose (ACROS Organics Geel, Belgium), 0.1% starch (Spectrum, New Brunswick, NJ), 2% gelatin (G Biosciences, St. Louis, MO) and 1% yeast extract (BD, Franklin Lakes, NJ) (sBHI). All cultures were incubated at 37°C. Anaerobic conditions were generated by the GasPak EZ Anaerobe Container System (BD, Franklin Lakes, NJ) system. Organisms requiring CO2 (5%) for growth, were cultured in CO2 incubator (VWR Symphony). Liquid cultures were incubated until visible turbidity was detected. All incubations were carried out at 37°C.
Organism |
Solid Media |
Liquid Media |
Culture conditions |
Gardnerella vaginalis |
Blood |
sBHI |
Anaerobic |
Atopobium vaginae |
Blood |
sBHI |
Anaerobic |
Lactobacillus crispatus |
MRS |
MRS |
5%CO2 |
Lactobacillus iners |
HBT |
sBHI |
5%CO2 |
Streptococcus agalactiae |
BHI |
BHI |
Aerobic |
Bacteroides fragilis |
Blood |
sBHI |
Anaerobic |
Prevotella bivia |
Blood |
sBHI |
Anaerobic |
Clostridium septicum |
Blood |
sBHI |
Anaerobic |
Fusobacterium nucleatum |
Blood |
sBHI |
Anaerobic |
Veillonella parvula |
Blood |
sBHI |
Anaerobic |
Enterococcus faecalis |
BHI |
BHI |
Aerobic |
Candida albicans |
SBA |
LB |
Aerobic |
Neisseria meningitidis |
BHI |
BHI |
5%CO2 |
Staphylococcus aureus |
BHI |
BHI |
Aerobic |
Pseudomonas aeruginosa |
LB |
LB |
Aerobic |
Escherichia coli |
LB |
LB |
Aerobic |
Table 1 Organisms cultivated and the appropriate media and growth conditions.
Blood agar - Brucella, CDC Anaerobe, or TSA II media were used
Bacterial enumeration and genomic DNA isolation
Once plank tonic bacterial growth had been achieved, an aliquot of each culture was removed to measure the Optical Density at 600nm (OD600). Liquid cultures were then serially diluted and plated onto appropriate solid media to enumerate the viable cells in each. Aliquots of each culture were also enumerated for total cells with a Z1 Particle Counter (Beckman-Coulter, Brea, CA). Aliquots of these cultures were used to isolate genomic DNA from each bacterial species via the DNeasy blood and tissue kit (Qiagen Valencia, CA). Isolated DNA was quantitated on a NanoDrop 8000 Spectrophotometer. A genome copy calculator from Thermo Scientific was used to ensure that 109 total copies/mL were used for each mock sample.
Clinical study participants
We undertook a retrospective diagnostic cross-sectional study with consecutive recruitment of all next-generation sequencing (NGS) vaginal swab specimens examined at American International Biotechnology LLC (AIBioTech, Richmond, VA) between July 1, 2014 and November 30, 2014. Vaginal swabs were collected from 270 women from community gynecology practices in the United States. All study participants consented to participate. All samples were de-identified to researchers involved in this study in order to maintain patient confidentiality.
DNA Extraction
Cervical and Vaginal swab specimens were collected and stored using the BD™ Universal Viral Transport System (Becton Dickinson, MD) according to the manufacturer’s instructions. Extraction of DNA was performed using the RTP Bacteria DNA Mini Kit (STRATEC Molecular, Berlin) according to the manufacturer’s instructions. Briefly, the sample is mixed and 400μl is transferred to the provided extraction tube. 400μl Re-suspension Buffer added, incubated for 10min at 37°C in a thermo mixer, then for 10min at 65°C in a thermo mixer, then for 5-10minutes at 95°C in a thermo mixer. 400μl Binding Buffer B6 is added, vortexed, and transferred to RTA-Spin Filter. The column is washed twice with the provided wash buffers. To eliminate residual ethanol, samples are centrifuged for 4min at maximum speed. Samples are eluted with 200μl of the provided elution buffer at 65°C. Samples are stored at 4°C for up to one month or stored at –20°C.
Semi-conductor based next generation sequencing
Universal primers were designed to amplify multiple regions of the 16S rRNA gene sequences. DNA libraries were generated using the Ion AmpliSeq™ Library Kit 2.0-96LV (Cat. no. 4480441). See Ion AmpliSeq™ Library Preparation (Publication Part Number MAN0006735 Revision A.0). Two separate primer pools were utilized in the initial amplification reaction. The pools were combined before proceeding with the FuPa reaction. Ion Xpress™ Barcode Adapters 1-16 Kit (Cat. No. 4471250) and Barcode Adapters 17-32 Kit (Cat. No. 4474009) were utilized for sample barcoding and adaptor ligation. AmPureAgencourt XP (Cat. No. A63882, Beckman) magnetic PCR cleanup beads were utilized as indicated in the Ion AmpliSeq™ Library Preparation User Guide. Sample libraries were normalized by Ion Library Equalizer™ Kit (Cat. no. 4482298), yielding a final sample library of 100μL at 100pM concentration. Emulsion PCR (emPCR) was performed on the Ion One Touch™ 2 Instrument (Cat. No. 4474778) as indicated in Ion PGM™ Template OT2 200 Kit (Publication Number MAN0007220. Revision 5.0). Reagent kit Ion PGM™ Template OT2 200Kit (Cat. no. 4480974). Briefly, normalized 100pM sample libraries were pooled and loaded with OT2 kit reagents with ISP beads. Samples were processed for enrichment on the Ion One Touch™ ES no later than 16hours post emPCR run completion. Enrichment of ISPs was achieved using Reagent kit Ion PGM™ Template OT2 200 Kit (Cat. no. 4480974) and DynaBeadsMyOne streptavidin C1 beads (cat. no. 65001, Life Technologies) according to the manufacturer's protocols (Ion PGM™ Template OT2 200 Kit User Guide). Ion Torrent Ion 316™ Chip Kit v2 (Cat. No. 4483188, Life Technologies) were prepared and loaded according to the manufacturer's recommendation (Ion PGM™ Sequencing 200Kit v2 (Publication Number MAN0007273 Revision 3.0). Chips were manually loaded with enriched ISPs with primed sequencing polymerase (provided in kit) using Rainin® pipette tips SR-L200F. The Ion Torrent PGM was run according to Ion Torrent 316 chip specifications, 500 flows, and use of 18 MΩ water system (multistage system including: Carbon tank, RO membrane, UV irradiation, post-filtering, deionization, and an Elga water polisher). Standard compressed nitrogen gas supplied to the PGM system.
Analysis of next generation sequencing data
A reference 16S rDNA non-redundant, database was compiled for organisms likely to be present in the vagina based on data from the Vaginal Microbiome Consortium.20 This resource is based upon an in-depth analysis of over 1000 participants. Full 16S sequences were compiled for over 1,300 bacteria and used for comparison of targeted sequencing data. A custom bioinformatics solution was created to process the data using PERL. Reads greater than 75 nucleotides in length were used to align to a manually curated USEARCH database of the organisms described.24 In order to be considered as detected, the organism had to exceed a threshold of 100 reads across multiple variable regions of the 16S rDNA molecule. Each detected organism was approximated by dividing the number of reads mapped to the organism by the total number of reads mapped to all bacteria. Appropriate controls were included in each sequencing run including a positive control composed of multiple bacteria in known proportions, an extraction blank and a non-template library blank. Acceptance of the sequencing results required that the positive control return each bacterium in the mix, and less than 7500 reads with no organism calls for both blanks.
Statistical analysis
All statistical analyses were performed using R Project for Statistical Computing (http://www.r-project.org).25 A Kruskal-Wallis non-parametric analysis of variance was used to compare distributions between groups. All box plots were created using R. Microsoft Excel was used to create the stacked bar plots in (Figure 4 & 6).
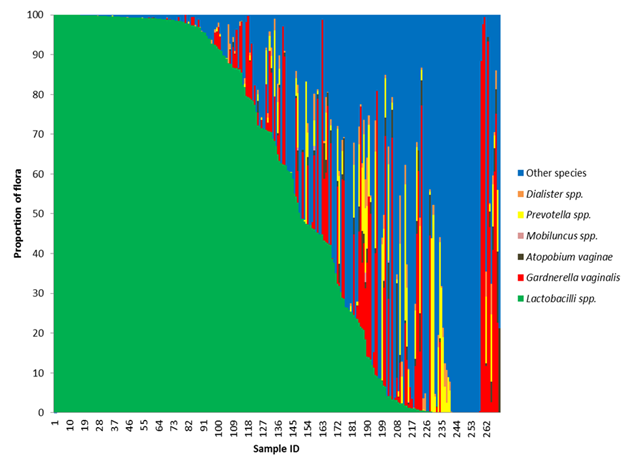
Figure 1 Loss of Lactobacilli species gives rise to non-resident bacteria.
The frequency of all Lactobacillus species, Gardnerella vaginalis, Atopobium vaginae, Megasphera species, Dialister species, Prevotella species were calculated for each participant in the study by dividing the sum of the reads for that organism by the total read count mapped to bacteria. If an organism was discovered that did not fit in the above referenced organisms, it was binned as other. A stacked bar graph was generated after sorting the participants by the frequency of Lactobacillus. As the frequency of Lactobacillus decreases, the frequency of the potential BV pathogens increases.
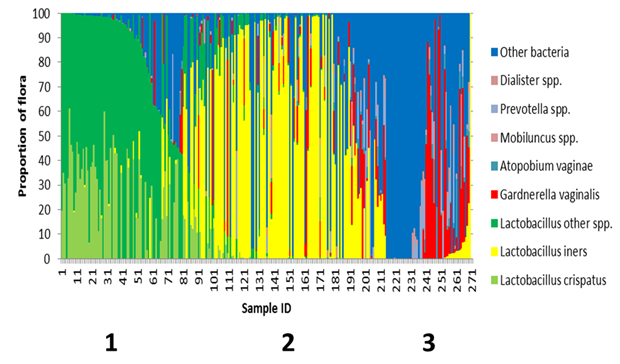
Figure 2 Speciation of Lactobacilli reveals a transitional flora.
The frequency of all Lactobacillus crispatus, Lactobacillus iners, and other hydrogen peroxide producing Lactobacillus species, Gardnerella vaginalis, Atopobium vaginae, Megasphera species, Dialister species and Prevotella species were calculated for each participant in the study by dividing the sum of the reads for that organism by the total read count mapped to bacteria. If an organism was discovered that did not fit in the above referenced organisms, it was binned as other. After sorting based on the proportion of Lactobacillus species, participants segregated into three distinct groups.
Mock communities
Species classification of organisms within a complex microbial community is inherently difficult due to the ubiquitous nature of the 16S rRNA molecule. Multiple mock communities were created to evaluate the ability to detect bacterial species commonly found in the vagina. The healthy vaginal flora is dominated by Lactobacillus crispatus. Multiple mock samples were created to test the ability to detect organisms found in vaginal samples in the presence of a higher number of L. crispatus genome copies (Table 2). With one exception, the organisms evaluated were positively classified at the species level. E. coli was incorrectly called as a Shigella species. Many of the organisms included in the mock community testing are important as they are suspected to be involved in the pathogenesis of bacterial vaginosis.
Lactobacillus crispatus genome copies |
Other organism |
Other bacteria genome copies |
Both Detected |
1.90 x 106 |
Lactobacillus iners |
1.90x105 |
Yes |
1.90 x 106 |
Gardnerella vaginalis |
1.90x105 |
Yes |
1.90 x 106 |
Fusobacterium nucleatum |
1.90x105 |
Yes |
1.90 x 106 |
Veillonella parvula |
1.90x105 |
Yes |
1.90 x 106 |
Streptococcus agalactiae |
1.90x105 |
Yes |
1.90 x 106 |
Prevotella bivia |
1.90x105 |
Yes |
1.90 x 106 |
Atopobium vaginae |
1.90x105 |
Yes |
1.90 x 106 |
Bacteroides fragilis |
1.90x105 |
Yes |
1.90 x 106 |
Clostridium septicum |
1.90x105 |
Yes |
1.90 x 106 |
Enterococcus faecalis |
1.90x105 |
Yes |
1.90 x 106 |
Neisseria meningitis |
1.90x105 |
Yes |
1.90 x 106 |
Staphylococcus aureus |
1.90x105 |
Yes |
1.90 x 106 |
Pseudomonas aeruginosa |
1.90x105 |
Yes |
1.90 x106 |
Escherichia coli |
1.90x105 |
No* |
Table 2 Mock Communities Containing Bacteria in Contrasting Proportions
* Escherichia coliwas incorrectly called as Shigella spp
The vagina is a complex environment which can be inhabited by multiple bacterial species as well as a number of fungal organisms. Vulvovaginal candidiasis occurs in 75% of women over 25 years of age. Candida albicans is the most common causative agent of yeast infections worldwide. It is not uncommon for a woman to have concomitant bacterial vaginosis and Vulvovaginal candidiasis. A clinical assay to diagnose BV must be able to accurately distinguish these syndromes from one another. To ensure that our assay could detect BV related organisms in the presence of C. albicans, three unique mock communities were created containing Candida albicans along with various bacterial species related to BV. Each mock sample contained a unique mixture of bacterial genomes and a varying number of C. albicans genome copies (Table 3B-D). Bacteria were amplified and sequenced in the presence of C. albicans (Table 3A), likewise C. albicans was amplified and sequenced in the presence of the bacterial components. Further, each bacterium in the Candidia mixes was correctly identified at the species level. Analysis of the mock communities demonstrated that this next generation sequencing approach was capable of correctly identifying members of a complex bacterial and fungal community to the species level and within a milieu of proportions that varied in several orders of magnitude. These mock communities established the validity of the assay.
Sample ID |
Bacteria genome copies |
Candida albicansgenome copies |
Bacteria Detected |
Candida albicans Detected |
CV1 |
5.0x 108 |
1.0 x 106 |
Yes |
Yes |
CV2 |
6.0 x 108 |
8.0x105 |
Yes |
Yes |
CV3 |
7.5 x108 |
5.0x105 |
Yes |
Yes |
Table 3 Mock Communities Containing Bacteria and Candida albicans.
Table 3A
Bacteria Expected in Mix CV1 |
Genome Copies |
Bacteria Found |
Lactobacillus crispatus |
100000 |
Yes |
Lactobacillus iners |
100000 |
Yes |
Streptococcus agalactiae |
400000 |
Yes |
Enterococcus faecalis |
100000 |
Yes |
Staphylococcus aureus |
200000 |
Yes |
Pseudomonas aeruginosa |
100000 |
Yes |
Table 3B
Bacteria Expected in Mix CV2 |
Genome Copies |
Bacteria Found |
Lactobacillus iners |
200000 |
Yes |
Gardnerella vaginalis |
300000 |
Yes |
Prevotella bivia |
600000 |
Yes |
Atopobium vaginae |
200000 |
Yes |
Bacteroides fragilis |
100000 |
Yes |
Neisseria gonorrhoeae |
100000 |
Yes |
Table 3C
Bacteria Expected in Mix CV3 |
Genome Copies |
Bacteria Found |
Fusobacterium nucleatum |
200000 |
Yes |
Veillonella parvula |
300000 |
Yes |
Staphylococcus aureus |
200000 |
Yes |
Table 3D
Clinical samples
To our knowledge, this is the first clinical application of a next generation sequencing assay for diagnosis of BV. This application has only recently become possible due to reductions in sequencing cost and increases in throughput. The cost to consumer of this assay is similar to that of other molecular diagnostic assays used to test for BV. In order for a clinical assay to provide clinically meaningful data, it must be returned to the physician in a meaningful period of time. Mean turnaround time for this assay was 3.7 days from sample receipt to final written report to clinicians. With this rapid turnaround, for the first time clinicians are able to receive clinically actionable information that shows a more comprehensive picture of the vaginal microbiome than any single target stand-alone test can provide. Armed with these results, clinicians can accurately assess the entirety of the vaginal flora.
After collection in the clinic, vaginal or cervical swabs (n= 270) were sent by overnight delivery to the lab for processing and DNA extracted within 24 hours of receipt. Average age of participants in this study was 36 years (SD= 14, min=18, max =90). The mean number of reads sequenced in each sample was 153, 545. In the samples collected, a large number of diverse organisms were found (Table 4). The most common potential pathogen was Gardnerella vaginalis found in ~ 65% of cases. Until recently, G. vaginalis was considered the sole causative agent of bacterial vaginosis. Current evidence challenges this hypothesis as G. vaginalis may be present in up to 60% of women that do not have symptoms of BV.26 In a paper published in 1955, Gardner and Dukes inoculated 13 healthy women with pure cultures of G. vaginalis and only one developed any symptoms associated with BV.27 It appears other factors are involved in the pathogenesis in at least some cases of BV. Analysis of the microbial community revealed that the number of unique bacterial genera significantly differs depending upon the presence or absence of Gardnerella vaginalis (Figure 3A). When G. vaginalis is present, there tended to be a larger number of bacterial genera present (µ=10.6 vs 4.7).
Organism |
Detected |
Not Detected |
Lactobacillus iners |
152 |
118 |
Gardnerella vaginalis |
106 |
164 |
Lactobacillus crispatus |
90 |
180 |
Atopobium vaginae |
66 |
204 |
BVAB2 |
50 |
220 |
Megasphaera spp. |
48 |
222 |
Prevotella bivia |
44 |
226 |
Sneathia sanguinegens |
39 |
231 |
BVAB1 |
27 |
243 |
BVAB3 |
16 |
254 |
Mobiluncus curtisii |
5 |
265 |
Mobiluncus mulieris |
4 |
266 |
Bacteroides fragilis |
1 |
269 |
Table 4 BV associated organisms
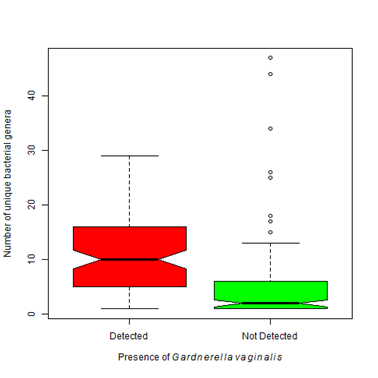
Figure 3 Number of bacterial genera detected varies depending upon organisms present.
3A. Gardnerella vaginalisis found most often when accompanied by an environment that contains a higher bacterial biodiversity. That is the number of detected bacterial genera significantly differs between patients with Gardnerella vaginalis and those that do not (Kruskal-Wallis Χ2 = 73.45, df=1, p-value < 0.0001).
Lactobacilli species tend to dominate the vagina of women without BV.28 An important component of a healthy vaginal flora is Lactobacillus crispatus .26 It is hypothesized that during the development of BV, the healthy Lactobacilli species, such as L. crispatus are displaced by other potentially more pathogenic agents. L. crispatus was found in 50% of our cases (Table 4). Unlike G. vaginalis, the presence of Lactobacillus crispatus was associated with a lower number of detected bacterial genera (µ=4.07 vs 7.0) (Figure 3B). Other Lactobacilli species are also components of the vaginal flora such as L. jensenii, L. gasseria, L. helveticus, L. gasseri and L. vaginalis.29 They produce hydrogen peroxide and lactic acid which are responsible for maintenance of an acidic environment and prevent colonization by some pathogens.30 Our results showed that when Gardnerella vaginalis was present, a lower proportion of healthy lactobacilli species were found indicating that healthy bacteria were lost as Gardnerella vaginalis colonized (Figure 4A). The presence of Lactobacillus crispatus was associated with a higher proportion of healthy lactobacilli species (60% vs 30%) (Figure 4B).
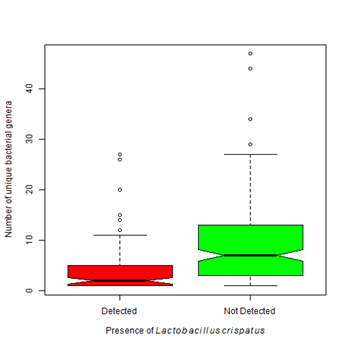
Figure 3B Vaginal samples colonized with Lactobacillus crispatusare associated with a lower bacterial biodiversity than those that do not contain this organism (Kruskal-Wallis Χ2 = 35.40, df=1, p-value < 0.0001).
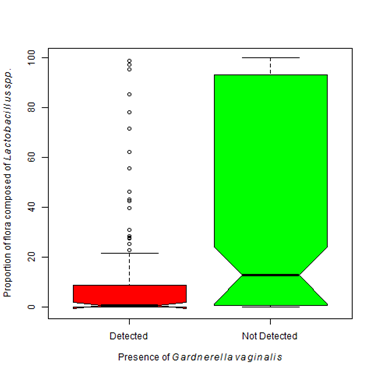
Figure 4 Healthy Lactobacillus species vary depending upon floral composition.
4A. The presence of Gardnerella vaginalis is associated with a lower proportion of Lactobacilli species excluding L. iners. The proportion of Lactobacilli species significantly differs with the presence of G. vaginalis (Kruskal-Wallis Χ2 = 26.29, df=1, p-value < 0.0001).
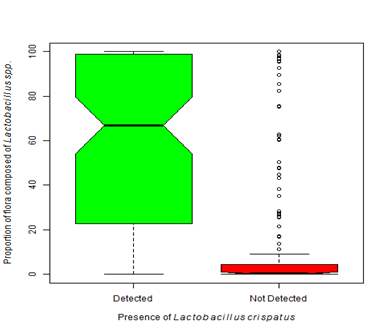
Figure 4B Dominance by one or more Lactobacilli species is considered normal. The presence of Lactobacillus crispatus is often described as protective. Our analysis shows that when Lactobacillus crispatus is present, the proportion of flora dominated by Lactobacillus species is significantly greater than when L. crispatus is not present (Kruskal-Wallis Χ2 = 107.39, df=1, p-value < 0.0001).
Other bacterial genera have been found in patients with bacterial vaginosis such as Megasphera, Atopobium, Sneathia, Prevotella, Bacteroides, Mobiluncus, Urea plasma and Mycoplasma. Our results showed a larger proportion of these BV-associated bacteria were found in samples containing G. vaginalis (Χ2 = 179.24, df=1, p-value < 0.0001). Nearly 33% of the total flora was composed of pathogenic agents when G. vaginalis was present compared to only 3% when absent (Figure 5A). The presence of Lactobacillus crispatus was associated with a lower proportion of reported BV associated organisms (<5% vs 21.4%) (Figure 5B).
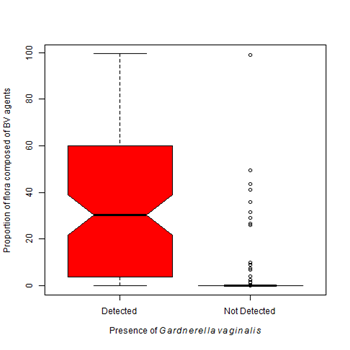
Figure 5 Proportion of pathogens varies with Gardnerella vaginalis and Lactobacillus crispatus.
5A. The presence of Gardnerella vaginalis is most often associated with a flora dominated by species associated with the development of bacteria vaginosis. The proportion of flora occupied by BV associated organisms significantly differs between an environment colonized with G. vaginalis and an environment free from G. vaginalis (Kruskal-Wallis Χ2 = 179.24, df=1, p-value < 0.0001).
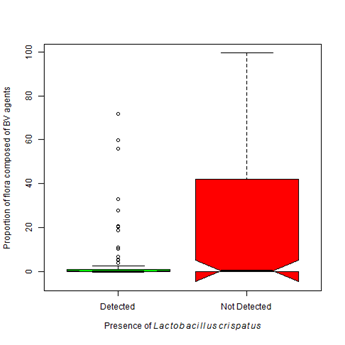
Figure 5B Proportion of pathogens varies with Gardnerella vaginalis and Lactobacillus crispatus.
5A. The presence of Gardnerella vaginalis is most often associated with a flora dominated by species associated with the development of bacteria vaginosis. The proportion of flora occupied by BV associated organisms significantly differs between an environment colonized with G. vaginalis and an environment free from G. vaginalis (Kruskal-Wallis Χ2 = 179.24, df=1, p-value < 0.0001).
A benefit of the next generation sequencing approach was the ability to quantify proportions of bacterial reads and compare samples to one another. A plot of the bacterial composition was generated for each participant. For this analysis, all Lactobacilli species were combined into a single group and organisms that are not typically associated with BV were grouped into other species (Figure 1). We found that in cases with a lower proportion of L. crispatus and other Lactobacilli species, there was a higher proportion of G. vaginalis and other potentially pathogenic species. Only a small number of patients (14) were colonized with a majority of G. vaginalis (ie. greater than 50%). The majority of the participants that tested positive for G. vaginalis also had other BV-associated organisms. There was a relatively high frequency of cases (n= 77) with a high proportion of other organisms not traditionally associated with BV; such as Enterococcus, Staphylococcus, Streptococcus, Clostridium and Aurantimonas. The mean number of bacterial genera detected within the other organism dominated group was 10.2. The high bacterial diversity and absence of healthy, protective Lactobacilli species was suggestive of bacterial vaginosis.
In (Figure 1), all Lactobacilli species were grouped into a single grouping. It is known that not all Lactobacilli offer the same protection as L. crispatus. Where L. crispatusis considered protective because of its ability to produce hydrogen peroxide, most strains of L. iners are non-H 2O2 producers, thus they are less protective than other Lactobacilli strains.31 Lactobacillus iners seems to be capable of surviving in diverse vaginal environments and in the face of multiple bacterial communities.26,32 These bacteria were found more often than any other organism in our study (Table 4). We found Lactobacillus iners in the presence of Lactobacillus crispatus and Gardnerella vaginalis (Figure 6A). Even though it can be found in the context of a normal flora, statistically it is more often found in higher proportions in the presence of Gardnerella vaginalis (Χ2 = 7.83, df=1, p-value =0.005). There was not a statistically significant difference in the proportion of L. iners found in patients colonized with L. crispatus as compared to those that are not (Figure 6B). L. iners may be found in a healthy, asymptomatic woman, or in a patient showing clear symptoms of vaginosis.33 Treatment of BV with metronidazole causes the proportion of L. iners to increase without a subsequent increase in L. crispatus.34
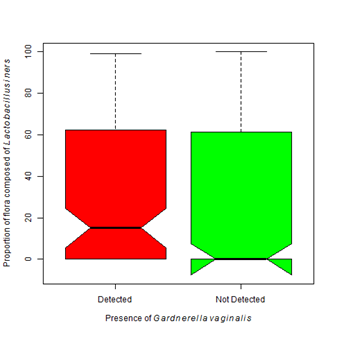
Figure 6 Colonization of Lactobacillus iners is more associated with Gardnerella vaginalis.
6A. The presence of Gardnerella vaginalis is associated with a higher proportion of colonization by Lactobacillus iners (Kruskal-Wallis Χ2 = 7.83, df=1, p-value =0.005).
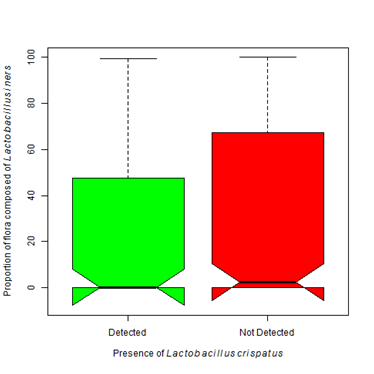
Figure 6B The presence of Lactobacillus iners does not statistically differ with the presence of Lactobacillus crispatus (Kruskal-Wallis Χ2 = 2.20, df=1, p-value =0.138).
Not all Lactobacilli species are equal, L. crispatus likely serves a protective role as it is a strong peroxide producer.31 The majority of women are colonized by more than one species of Lactobacilli. A second bacterial composition plot was generated that broke Lactobacilli into three groups; L. crispatus, L. iners and other hydrogen producing Lactobacillus species (Figure 2). In this figure, there exist three distinct groups that segregate largely based on Lactobacilli. Group 1 participants possessed a flora dominated by healthy Lactobacilli species and the majority of these participants were colonized with L. crispatus. As the proportion of hydrogen producing Lactobacilli decrease, the proportion of L. iners increased. Group 2 was L. iners dominant. There were a total of 152 participants that were colonized with some degree of L. iners. Of those participants, 50% were colonized with G. vaginalis. The flora of these participants was likely a transitional/ intermediate flora.35 The third group of participants was more likely to show symptoms of BV based on the microbial composition. The majority of this group was colonized with some degree of G. vaginalis.
The results of this study showed that BV was likely present when there were few healthy Lactobacilli species present to protect the vagina from non-resident microorganisms and the sum proportion of non-resident species was greater than the proportion of L. iners. Of the 270 participants in this study, 105 participants were predicted to be BV positive and the remainder predicted as BV negative. The three most commonly found organisms in samples predicted to be BV positive were Gardnerella vaginalis, Lactobacillus iners and Atopobium vaginae (Table 5). Over 60% (n=65) of participants predicted BV positive were infected with Gardnerella vaginalis and Atopobium vaginae. Other studies have also shown that Atopobium vaginae rarely occurs in isolation, it is nearly always found in association with Gardnerella .36 Co-infection with G. vaginalis and A. vaginae is more likely to occur in women with recurrent BV infection. G. vaginalis was found in approximately 10% of women that were likely not suffering from BV symptoms. The majority of their bacterial flora was composed of healthy Lactobacilli (Figure 2). Organisms commonly associated with BV are rarely found in patients predicted to BV negative.32
Organism |
BV Positive |
BV Negative |
Lactobacillus iners |
67 |
85 |
Gardnerella vaginalis |
87 |
19 |
Lactobacillus crispatus |
12 |
78 |
Atopobium vaginae |
65 |
1 |
BVAB2 |
50 |
0 |
Megasphaera spp. |
48 |
0 |
Prevotella bivia |
39 |
5 |
Sneathia sanguinegens |
39 |
0 |
BVAB1 |
27 |
0 |
BVAB2 |
16 |
0 |
Mobiluncus curtisii |
5 |
0 |
Mobiluncus mulieris |
4 |
0 |
Bacteroides fragilis |
1 |
0 |
Table 5 Comparison of organisms after applying model for BV prediction
This study is one of the first studies to bring next generation sequencing of vaginal samples to community practices across the United States. This study shows that an NGS based, metagenomic assay can be delivered to clinical practices across the United States and deliver results within a time frame that can guide clinical treatment. This approach is superior to other diagnostic assays that often only look for the presence of single bacteria like Gardnerella vaginalis. NGS assays provide more comprehensive data that reveal a more complete picture of the distribution of bacteria giving clinicians a more complete understanding of their patient’s vaginal flora. This approach is unbiased and does not rely on the incomplete knowledge of the etiology of BV. Our results showed that there existed three vaginal profiles that largely segregate based on the presence of facultative anaerobic organisms and the absence of healthy Lactobacilli. The first group was largely dominated by Lactobacilli species and was relatively homogenous. The second group was dominated by Lactobacillus iners and was more closely related to BV than the homogenous flora of group 1. Conversely, the flora of the third group was not dominated by any single organism but rather contained a mixture of organisms.
The authors of this manuscript acknowledge the efforts of all clinicians that referred participants for this study. We wish to thank Dr. Courtney A. Hunt for her efforts and her gracious review of the manuscript. We also think all employees of AIBioTech for their work on development of this project.
The authors of this study are employed by American International Biotechnology, which is a comprehensive contract research organization that offers clinical next generation sequencing services.

©2015 Budd, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.