MOJ
eISSN: 2374-6912


Research Article Volume 3 Issue 3
1Department of Otolaryngology, Head and Neck Surgery, Hainan Branch of Chinese PLA General Hospital, China
2Center for Biotechnology and Genomic Medicine, Medical College of Georgia, USA
3Department of Otolaryngology, Head and Neck Surgery, First Affiliated Hospital of Anhui Medical University, China
Correspondence: Minbo Liu, Department of Otolaryngology, Head and Neck Surgery, Hainan Branch of Chinese PLA General Hospital, Sanya, Hainan Province 572000, China
Co-correspondence: Liyong Zhang, Center for Biotechnology and Genomic Medicine,Medical College of Georgia, Augusta, GA 30912, USA
Received: May 05, 2017 | Published: August 11, 2017
Citation: Han M, Liu Z, Wang F, et al. Identification of differentially expressed proteins from prostate cancer cell lines by fluorescence 2-D difference gel electrophoresis (2-D DIGE). MOJ Cell Sci Rep. 2016;3(3):81-88. DOI: 10.15406/mojcsr.2016.03.00059
Prostate cancer (PCa) is the most common cancer diagnosis and the second leading cause of cancer-related deaths in males worldwide. Prostate-specific antigen (PSA) is the only biomarker widely utilized in diagnosis and management of PCa patients. However, PSA lacks sensitivity and specificity for diagnosis, which leads to false-negative and false-positive test results. PSA cannot differentiate indolent from aggressive prostate disease, which leads to over-treatment of many patients with associated side-effects. Furthermore, PSA is unable to identify tumors which are likely to become unresponsive to treatment at an early stage. Thus, there is an urgent need for identification of biomarkers which will improve the diagnosis and management of PCa. In this study, to explore the specific proteins for PCa, fluorescence 2-D Difference Gel Electrophoresis (2-D DIGE) was performed in 3 prostate cancer cell lines (LNCaP, PC-3, and DU145) with the comparison of normal prostate cells (RWPE-1). The separated proteins with a fold difference greater than two and presence in all both normal and cancer cells were identified by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF/TOF). Expression profiling between prostate cancer cell lines and normal prostate cells revealed 33 protein candidates with expression difference (P<0.05) and over 2-fold change. The expression levels of peroxiredoxin 3 (PRDX3), keratin 8 (KRT8), and heat shock protein 60 (HSP60) were verified with Western blot in the normal epithelial cells RWPE-1 and prostate cancer cell lines LNCaP, PC-3, DU145 as well 25 pairs of patient-matched prostate tissues. The proteins are aberrantly expressed in human prostate cancer suggesting their implication in the development and progression of this malignancy.
Keywords:prostate cancer, 2-D DIGE, MALDI-TOF
Prostate cancer (PCa) is the most commonly diagnosed malignancy and the second most common largest cancer in males worldwide.1 Approximately 2,30,000 patients were diagnosed prostate cancer in the US only in 2012. Nearly 40 000 persons were kill by the disease and its complications in the US annually which makes it the second leading cause of cancer death.2,3 Prostate specific antigen (PSA) is secreted by the epithelial cells of the prostate and by their derivatives, the prostate cancer cells. It leaks into the circulation and serum PSA is used as a diagnostic marker to monitor the progress of prostate cancer during treatment. However, PSA lacks sensitivity and specificity when it is used to screen men for prostate cancer.4 Over the past decade, large concerted efforts involving proteomic, transcriptomic, genomic, and metabolomics strategies, have been aimed to identify reliable biomarkers for PCa from a wide range of samples including urine, serum, tissues, exosomes, and cell lines.3,5-17 Although a number of candidate biomarkers have been identified,6-8,13,18-23 these biomarkers have to be clinically validated prior to routine use.15,16,22,24-26
2-D electrophoresis (2-DE) is a high-resolution technique that separates proteins based on their isoelectric point (pI) and molecular weight. Conventional 2-DE with Coomassie™ Blue or silver staining combined with image analysis is limited by the reproducibility, sensitivity and dynamic range of detection method.20,27,28 Manual comparison of gels contributes significantly to the inaccuracy, subjectivity, and tedium of proteome analysis.20 Difference gel electrophoresis (DIGE) is a technique whereby protein samples are labelled with different fluorescent dyes so they can be mixed together, co-separated, and visualized on a single 2-D gel for subsequent analysis.20 In addition, improvements in protein extraction, solubilization, labelling, separation, analysis and bioinformatics tools, have led to leaps in the identification of candidate disease biomarkers. Figure 1A shows the key stages of the DIGE method when using two protein extracts, each labelled with different fluorescent dyes.
In this study, three prostate cancer cell lines (LNCaP, PC-3, and DU145) and normal prostate cells (RWPE-1) were subjected to fluorescence 2-D Difference Gel Electrophoresis (2-D DIGE). The separated proteins with a fold difference greater than two and presence in all both normal and cancer cells were identified by matrix-assisted laser desorption/ionization (MALDI) TOF/TOF tandem mass spectrometry. These proteins aberrantly expressed in human prostate cancer suggested their implication in the development and progression of this malignancy.
In this study, three prostate cancer cell lines (LNCaP, PC-3, and DU145) and normal prostate cells (RWPE-1) were subjected to fluorescence 2-D Difference Gel Electrophoresis (2-D DIGE). The separated proteins with a fold difference greater than two and presence in all both normal and cancer cells were identified by matrix-assisted laser desorption/ionization (MALDI) TOF/TOF tandem mass spectrometry. These proteins aberrantly expressed in human prostate cancer suggested their implication in the development and progression of this malignancy.
Cell lines and prostate tissues
Normal adult human prostate epithelial cells RWPE-1 and prostate cancer cell lines DU145, LNCaP, PC-3 were purchased from American Type Culture Collection (ATCC, Manassas, VA). All tumor cell lines were maintained according to the recommended culture conditions provided by ATCC supplemented with 100μg/ml streptomycin and 100 μg/ml penicillin. Prostate tumor tissues and adjacent normal tissues used for this study were obtained at the time of diagnosis of prostate cancer. Written informed consent was procured from these subjects. The study was approved by the institutional Review Board of the Medical College of Georgia.
Reagents and antibodies
Electrophoresis reagents including acrylamide solution (40 %), N, N-methylenebis acryl amide, N, N, N’, N’-tetramethylethylenediamine, tris base, glycine, SDS, DTT, CHAPS, immobilized pH gradient strips, Pharmalyte, IPG buffer, IPG cover fluid, protein marker were from Amersham Pharmacia Biotechnology Inc. (Uppsala, Sweden); Iodoacetamide was from Acros (New Jersey, USA); Sequence grade Trypsin was from Washington Biochemical Corporation; Trifluoroacetic acid (TFA), CyDye™ DIGE Fluors (Cy2, Cy3, and Cy5), Urea and thiourea were bought from Fluka Chemical Corp. (Milwaukee, WI). CHAPS was obtained from Sigma. DTT was purchased from Fisher. All other reagents were of analytical grade.
Specific antibodies peroxiredoxin 3, keratin 8, and HSP60 were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). Antibody α-tubulin was from Sigma-Aldrich (St. Louis, MO). Goat anti-rabbit and goat anti-mouse HRP-conjugated secondary antibodies were from Promega (Madison, WI).
Sample preparation for 2-D DIGE
Phenol extraction of proteins was applied to prepare 2-DE samples as described previously.29-32 Briefly, cells were harvested by trypsin digestion when they reached 80-90% confluence, washed three times with PBS, and centrifuged at 400 × g for 5 min. Cell pellets were re-suspended in extraction buffer (500 mM Tris-HCl, 50 mM EDTA, 700 mM sucrose, 100 mM KCl, pH 8.0). This buffer can be stored for a week at 4°C. Two percent of β-mercaptoethanol and 1 mM phenylmethylsulfonyl fluoride (PMSF) were added just before extraction. After phenol extraction, proteins were precipitated with ammonium acetate in methanol. The pelleted proteins were then re-suspended in isoelectric focusing (IEF) buffer (9 M urea, 4% CHAPS, 0.5% Triton X-100, 20 mM DTT, 1.2% Pharmalytes), and the protein concentration was measured by the Bradford assay (Bio-Rad).
2-D DIGE and image analysis
According to the manufacture’s instructions, equal amount of proteins from each sample was labelled with fluorescent dyes.33 Briefly, 50 μg of lysates from each cell lines were labelled with 400 pM of CyDye DIGE Fluor minimal dyes freshly dissolved in anhydrous dimethyl formamide (DMF). The labelling mixture was incubated on ice in the dark for 30 min, and the reaction was terminated by addition of 10 nM of lysine. Two labelled samples were mixed with rehydration buffer (7 M urea, 2 M thiourea, 2% CHAPS, 50 mM DTT, 1% Pharmalyte) before being applied to an 18-cm immobilized pH gradient strip for overnight rehydration.
The 2-DE procedures were performed as described in our previous work (7) with modifications (20). Briefly, the first electrophoresis, isoelectric focusing (IEF), was carried out on an IPGphor isoelectric focusing (IEF) system (Amersham Pharmacia, San Francisco, CA) after the IPG strips (18 cm, 3–10 nonlinear pH gradient; GE Healthcare, Piscataway, NJ) were rehydrated with 250 μl of rehydration buffer. After IEF, the IPG gel strips were immediately equilibrated in 10 ml of equilibration buffer (50 mM Tris–HCl, pH 8.8 , 6 M urea, 30% glycerol, 2% SDS) containing 20 mM DTT for 15 min at room temperature and alkylated with 2.5% (w/v) iodoacetamide in 10 ml of equilibration buffer for another 15 min at room temperature. The strips were transferred onto vertical 13% SDS-PAGE gels and held in with 0.5% agarose dissolved in SDS/Tris running buffer. The electrophoresis were executed overnight at constant power (2.5 W/gel for 40 min and 15 W/gel for 6 hours) using Hoefer SE 600 Standard Vertical Electrophoresis Unit.
Gel images for analysis were obtained using Typhoon 9400 Imager using the following settings: Cy2 (488 nm excitation laser and 520BP40 emission filter); Cy3 (532 nm excitation laser and 580BP30 emission filter); and Cy5 (633 nm excitation laser and 670BP30 emission filter), and processed using DeCyder Differential Analysis Software. The spots on the gel were co-detected automatically as 2-D DIGE image pairs, which intrinsically link a sample to its in-gel standard. Matching between gels was performed utilizing the in-gel standard from each image pair. The experimental set-up and relationship between samples were assigned in DeCyder software. A student’s t-test was performed for every matched spot-set, comparing the average and standard deviation of protein abundance for a given spot between the control and experimental groups. Only those spots with differential expression (P<0.05) and over 2-fold changes in volume after normalization between the two populations were defined as altered.
Mass spectrometric analysis
For mass spectrometry analysis a preparative gel was run. The separated proteins in SDS-PAGE gels were visualized by SYPRO Ruby solution (Molecular Probes). The spots with a fold difference greater than two and presence on all both normal and cancer cell gels were excised as 2 mm-diameter plugs and delivered into 96-well microplates. The SYPRO Ruby stained gel was de-stained in 10% ethanol and 6% acetic acid, in-gel digested, and extracted as described previously (7,20). The dried peptides were dissolved in 0.5 % TFA. With saturated CHCA in 0.1 % TFA/50 % acetic acid as matrix, the peptide samples were analyzed by M@LDI R (Micromass, Manchester, UK). Mass spectra were internally calibrated using trypsin peaks. The peptide masses were searched against the protein databases of Swiss-prot/trEMBL (http://www.expasy.ch/tools/peptident.html) and Mascot (http://www.matrixscience.com/). One missed cleavage per peptide was allowed and an initial mass tolerance of 50 ppm was used in all searches. The proteins with more than 4 matched peptides were thought significant. The experimental procedures are illustrated as a flow chart and are presented in Figure 1B.
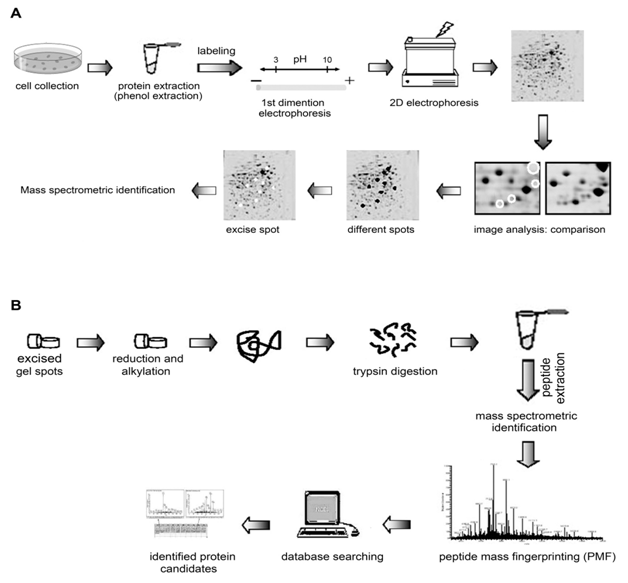
Figure 1 The schematic diagrams of the experimental strategy.
Western blot analysis
Cell pellets or tissues were lysed in lysis buffer (50 mM Tris-Cl, pH 7.5, 250 mM NaCl, 1 mM EDTA, 0.1% Triton X-100, and 1 × protein inhibitor cocktail). The protein concentrations were determined using the Bio-rad Protein Assay kit (Bio-Rad, Hercules, CA). Fifteen micrograms of proteins were separated and transferred to nitrocellulose membranes. After blocking in 5% milk for 1 hour at room temperature, the membranes were incubated with the appropriate primary antibody at 4°C overnight. After washing with TBST, the membranes were incubated with secondary antibody at room temperature for 1 hour. Proteins were detected with the enhanced chemiluminescence (ECL) kit (GE Healthcare, Piscataway, NJ). α-tubulin (Sigma) was used as loading control.
2D separation of proteins
Whole cell lysates from normal prostate epithelial cells RWPE-1, and three prostate cancer cell lines LNCaP, DU145, and PC-3 were produced and precipitated by phenol extraction,29-32 which eliminated lipids, nucleotides, and salts to improve the resolution of 2D gel analysis. The precipitated proteins from both normal cells and cancer cells were labeled with fluorescent dye Cy2, Cy3, or Cy5, respectively. And the resulting two pools of proteins were mixed and subjected to isoelectric focusing, followed by 13% SDS-PAGE in the 2nd dimension. 2D gel images of normal cells and cancer cells were generated using Typhoon 9400 Imager. A typical analytical 2-D DIGE experiment was shown in Figure 2.
All spots with a fold difference greater than two were considered for further analysis. Spots that appeared on all both normal and cancer cell gels were selected for analysis using MALDI TOF/TOF.
The changes of protein expression patterns between normal cells and cancer cells
Comparisons of protein expression in 2D images were carried out using DeCyder Differential Analysis Software. A student’s t-test was performed for every matched spot-set to compare the average and standard deviation of protein abundance for a given spot between the control and experimental groups. Only those spots with differential expression (P<0.05) and over 2-fold changes in volume after normalization between the two populations were defined as altered (Table 1-3).
Comparison of protein expression patterns between normal cells and cancer cells and SYPRO Ruby-stained gel images
To evaluate the global changes of protein pattern caused by dye labeling, the same gel was also stained with SYPRO Ruby. The overall protein-staining patterns between the SYPRO Ruby image and the dye images were very similar. However, there are subtle differences between the SYPRO Ruby image and the dye-labeled images, which could be caused by two major factors:
Protein identification by MALDI TOF/TOF mass spectrometry
Because the labeled protein has a molecular mass ~0.5 kDa higher than the unmodified protein, and the minimum number of molecules of each protein were labeled, the unlabeled protein spot rather than the labeled protein spot was excised for mass spectrometry analysis. The protein of interest was defined from the dye images, and its corresponding spot in the SYPRO Ruby image was matched. The protein spot was excised manually and subjected to in-gel digestion and mass spectrometer. The machine determined the molecular masses of tryptic peptides eluted from the HPLC column and their fragments, which are used to identify the protein by matching to known protein sequences existing in the protein databases of Swiss-prot/trEMBL (http://www.expasy.ch/tools/peptident.html) and Mascot (http://www.matrixscience.com/). The identified proteins were shown in Table 1-3
Gel Spot |
Differential Expression |
Protein Name |
Accession # |
F9/G3 |
10.36 |
Triosephosphate isomerase 1 [Homo sapiens] |
gi|17389815 |
D10/F4 |
7.31 |
Biliverdin reductase B (flavin reductase (NADPH)) |
gi|32891807 |
G10/A5 |
6.5 |
Neuropolypeptide h3 [Homo sapiens] |
gi|4261934 |
G10/A5 |
6.5 |
Prostatic binding protein [Homo sapiens] |
gi|4505621 |
G10/A5 |
6.5 |
Sarcoma antigen NY-SAR-41 (NY-SAR-41) [Homo sapiens] |
gi|56204569 |
B10/E4 |
4 |
Peroxiredoxin 3 isoform b [Homo sapiens] |
gi|32483377 |
E10/H4 |
4.21 |
Peroxiredoxin 2 isoform b [Homo sapiens] |
gi|33188452 |
E10/H4 |
4.21 |
Thiol-specific antioxidant protein [Homo sapiens] |
gi|438069 |
E10/H4 |
4.21 |
Peroxiredoxin 2, isoform a [Homo sapiens] |
gi|24659879 |
B9/G3 |
3.03 |
IDH1 protein [Homo sapiens] |
gi|15277488 |
B9/G3 |
3.03 |
NADP-dependent isocitrate dehydrogenase [Homo |
gi|3641398 |
C10/D4 |
2.97 |
Zinc finger protein ZNF224 [Homo sapiens] |
gi|6715532 |
F10/G4 |
2.93 |
Peroxiredoxin 1 [Homo sapiens] |
gi|55959887 |
G9/H3 |
2.69 |
Peroxiredoxin 6 [Homo sapiens] |
gi|56204402 |
D9/F3 |
2.27 |
X-Ray Crystal Structure Of The Human Galectin-3 |
gi|3402185 |
C9/H3 |
-3.5 |
hnRNP 2H9B [Homo sapiens] |
gi|7739445 |
Table 1 Protein identification of 2D gel spots from the comparison of normal prostate cell line with prostate cancer cell line LNCaP by MALDI-TOF/TOF
Gel Spot |
Differential Expression |
Protein Name |
Accession # |
B11/D5 |
2.39 |
Chain A, Arh-I, An AngiogeninRNASE A CHIMERA |
gi|23200076 |
B12/D6 |
3.5 |
Peroxiredoxin 6 [Homo sapiens] |
gi|56204402 |
C11/E5 |
3.19 |
KRT8 protein [Homo sapiens] |
gi|14198278 |
H10/B5 |
2.94 |
vinculin [Homo sapiens] |
gi|57162629 |
E11/G5 |
2.88 |
Keratin 19 [Homo sapiens] |
gi|24234699 |
D11/F5 |
2.23 |
Adenosine kinase [Homo sapiens] |
gi|30582415 |
G11/A6 |
-2.21 |
NUP107 protein [Homo sapiens] |
gi|34189309 |
Table 2 Protein identification of 2D gel spots from the comparison of normal prostate cell line with prostate cancer cell line PC-3 by MALDI TOF/TOF
Gel Spot |
Differential Expression |
Protein Name |
Accession # |
H8/B3 |
5.41 |
MRPL17 [Homo sapiens] |
gi|48146471 |
G8/A3 |
3.56 |
tumor necrosis factor, alpha-induced protein 3 |
gi|55664817 |
F6/B1 |
-2.87 |
KHSRP protein [Homo sapiens] |
gi|54648253 |
E6/A1 |
-2.95 |
KHSRP protein [Homo sapiens] |
gi|54648253 |
A7/D1 |
-3.35 |
KIAA0471 protein [Homo sapiens] |
gi|40788271 |
F8/H2 |
-3.46 |
CLE7 [Homo sapiens] |
gi|33150598 |
D7/F1 |
-3.88 |
Chain A, Complex Of The Kh3 And Kh4 Domains Of |
gi|20150334 |
H7/A2 |
-4.04 |
chaperonin 60, Hsp60 [Homo sapiens] |
gi|6996447 |
A8/D2 |
-4.37 |
IMP (inosine monophosphate) dehydrogenase 2 |
gi|13543973 |
D8/E2 |
-4.69 |
hnRNP 2H9B [Homo sapiens] |
gi|7739445 |
E7/H1 |
-7.49 |
FUBP1 protein [Homo sapiens] |
gi|16878077 |
Table 3 Protein identification of 2D gel spots from the comparison of normal prostate cell line with prostate cancer cell line DU145 by MALDI TOF/TOF
Validation of differentially expressed proteins
Three differentially expressed protein candidates peroxiredoxin 3 (PRDX3), keratin 8 (KRT8), and heat shock protein 60 (HSP60) were used to confirm the 2D DIGE results by Western blotting. The results indicated that compared with the normal epithelial cells RWPE-1, PRDX3 and KRT8 were overexpressed in the three prostate cancer cell lines LNCaP, PC-3, and DU145 while HSP60 was down-regulated in the three cancer cell lines (Figure 3A). As shown in Figure 3B, comparing their adjacent normal counterparts, the expression levels of PRDX3 and KRT8 were increased in 80% (20/25) and 88% (22/25) of 25 pairs of patient-matched prostate cancer tissues, respectively, whereas the expression levels of HSP60 were decreased in 84% (21/25) of the 25 paired cancer tissues. These results were consistent with those obtained from the 2D DIGE analysis.
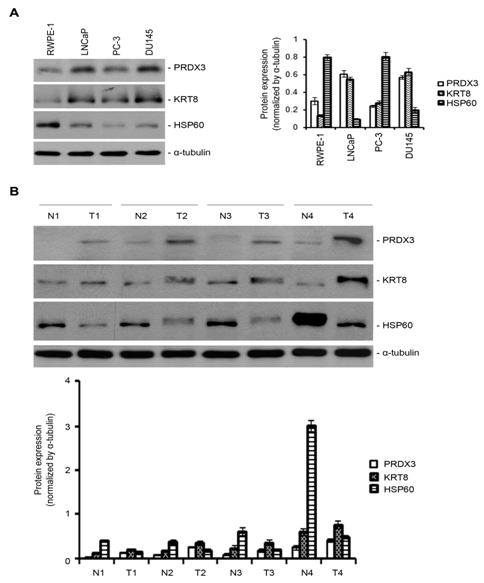
Figure 3 Validation of differentially expressed proteins by Western blotting.
Despite great advances achieved in proteomics, transcriptomics, genomics, metabolomics, molecular medicine, and translational research in prostate cancer,3,5–8 PCa remains an extremely frequent cancer and a common cause of cancer deaths in Western males. Obviously, early detection, targeted therapy and post-treatment surveillance are vital to fight this disease. Cancer biomarkers can be very helpful for early detection, prognosis prediction, and assessment of therapeutic effects. Currently, diagnostic tests such as PSA levels are not sensitive enough to determine whether a PCa is aggressive and likely to metastasize. Cancer cells are also known to influence the behavior and protein expression of the normal cells surrounding them. This might play a role in metastasis and aggressive invasion into normal tissue. Given the heterogeneity of PCa tissues, we cultured prostate cell lines ranging from normal adult human prostate epithelial cells RWPE-1, to hormone-dependent LNCaP (a less aggressive phenotype), and hormone-independent DU145, PC-3 (a highly aggressive phenotype) for the identification of potential biomarkers.34
The 2-DE is one of the powerful tools for separation of proteins from various clinical samples,7,35 but conventional 2-DE and image analysis is limited by reproducibility, sensitivity and detection methods.20,27,28 Manual comparison also contributes significantly to the inaccuracy of proteome analysis. Therefore, accurate measurement of differences in protein expression remains one of the most important aspects of proteomics. The reproducibility of conventional 2-DE can be improved dramatically by difference in-gel electrophoresis (DIGE).20,27 Furthermore, phenol extraction of proteins also allows efficient protein recovery and removes non-protein components such as lipids, salt, to enhance the resolution of 2-DE.29–32 The precipitated proteins from both normal cells and cancer cells were subsequently labelled with fluorescent dye Cy2, Cy3, or Cy5, respectively.33 The labeling conditions ensured that less than 5% of molecules of each protein were labeled. These dye molecules have molecular weights of approximately 0.5kDa and are mass- and charge-matched to give equivalent migration of the labelled proteins.20,27 The dyes are also positively charged to compensate for the positive charge that is lost during conjugation of the dye to lysine. Thus, the unlabeled protein spot rather than the labeled protein spot was excised from the preparative gel stained with SYPRO for mass spectrometry analysis.7 This analysis identified 16 spots with expression difference (P<0.05) and over 2-fold changes in volume after normalization between normal cells RWPE-1 and LNCaP cancer cells (Table 1), while comparing with the normal cells RWPE-1, 7 and 11 spots were found in PC-3 and DU145, respectively (Table 2 & 3).
Expression profiling between prostate cancer cell lines and normal prostate cells identified 33 protein candidates with expression difference (P<0.05) and over 2-fold changes in volume after normalization. As parallel experiments to identify the potential protein targets for the differentially expressed miRNAs for prostate cancer (2), peroxiredoxin 3 (PRDX3), keratin 8 (KRT8), and heat shock protein 60 were suggested as potential protein targets for the differentially expressed miRNAs by searching Target Scan database. The expression levels of these three protein candidates were on the priority to be confirmed by Western blot in the normal epithelial cells RWPE-1 and prostate cancer cell lines LNCaP, PC-3, DU145 as well 25 pairs of patient-matched prostate tissues.
These results were correlated with those obtained from the 2D DIGE analysis very well. PRDX3, originally identified from murine erythroleukemia, is a mitochondrial member of the anti-oxidant family of thioredoxin peroxidises.36 It is induced by oxidants in the cardiovascular system and is thought to play a role in the anti-oxidant defense system and homeostasis within the mitochondria.36 PRDX3 is also a c-myc target gene, and its expression is indispensable for neoplastic transformation by c-myc.36,37 It has been demonstrated that serum PRDX3 can be a valuable biomarker for the diagnosis and assessment of hepatocellular carcinoma.38 PRDX3 also protects MCF-7 breast cancer cells against doxorubicin-mediated toxicity.39 In addition, PRDX3 is up-regulated in prostate cancer and promotes cancer cell survival by defending cells against the damages incurred by oxidative stress.19,40 KRT8, also known as cytokeratin-8 (CK-8) or keratin-8 (K8), can be used to distinguish lobular carcinoma from ductal carcinoma of the breast.41 CAM 5.2, an antibody reacting with both CK8 and CK18, is employed in immunohistochemistry to define certain forms of cancer.42
In normal tissue, it reacts mainly with secretory epithelia instead of squamous epithelium. However, it reacts with a range of malignant cells including those derived from secretory epithelia, and some squamous carcinoma, such as spindle cell carcinoma.42 It is very helpful in the identification of microscopic metastases of breast carcinoma in lymph nodes,41 and in differentiation of Paget’s disease from malignant melanoma.43 It also reacts with neuroendocrine tumors.44 It is frequently used together with keratin 18 and keratin 19 to differentiate cells of epithelial origin from hematopoietic cells in tests that enumerate circulating tumor cells in blood.45 In prostate cancer cell lines, KRT8 also presents on the cell surface of transformed prostate cancer cells.46 The expression levels of KRT8 are high on the surfaces of DU145 and PC-3 cells but relatively low on the surfaces of LNCaP cells, and RWPE-1 cells, indicating it might be an important marker to distinguish prostate cancer from progression.46
HSP60, a mitochondrial chaperonin, is typically responsible for transportation and refolding of proteins from the cytoplasm into the mitochondrial matrix.47 Further studies have implicated HSP60 in stress response, diabetes,48–50 cancer51–53 and certain types of immunological disorders.54 HSP60 has been shown to affect apoptosis in tumor cells.52,53,55 The expression of HSP60 is increased in poorly differentiated prostate cancers,56 and its expression levels are correlated very well with clinical parameters.56 Thus, its changes in expression level have been shown to be useful for diagnostic and prognostic purposes.52,57 Identification of such kind of biomarkers could be of relevance in the clinical management of prostate cancer.
In summary, we have successfully executed the 2D DIGE proteomic profiling in context of prostate cancer. The most prominent spots were successfully identified and several protein candidates were validated in prostate tissues. The exact role of these protein candidates and their signal cascades in prostate cancer remains to be investigated in the future. The clinical utility of these protein candidates needs to be addressed. However, we believe that our data present the foundation for future studies in prostate cancer.
None.
The author declares no conflict of interest.

©2017 Han, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.