MOJ
eISSN: 2374-6912


Research Article Volume 4 Issue 5
Department of Botany, Mycology and Plant Pathology Laboratory, Dr Harisingh Gour University, India
Correspondence: Rafiq Ahmad Dar, Department of Botany, Mycology and Plant Pathology Laboratory, Dr Harisingh Gour University, India
Received: October 30, 2017 | Published: November 16, 2017
Citation: Dar RA, Rai AN. Origin of Alternaria kashmeriana causing canker stains on native Platanus orientalis kashmeriana - a first report from the Asia. MOJ Cell Sci Rep. 2017;4(5):132-136. DOI: 10.15406/mojcsr.2017.04.00099
In mid-summer 2015, during the mycological explorations to the northern India (Kashmir), canker stain symptoms in alarming proportions were observed on beneath of the leaf surfaces in 21 differentially located Platanus orientalis kashmeriana populations similar to what has been reported earlier in United States and Europe. Genetic interpretations of these isolates, using ITS region of rDNA as unique DNA barcoding site for molecular identification showed the evolution of new fungal species, Alternaria kashmeriana sp.nov. From closely related species under geographically distinct conditions encountering 0.5 amount of genetic change in ITS region of rDNA. It is hard to differentiate the Alternaria Kashmeriana sp.nov and from the rest known Alternaria species on the phenotypic bases but reflects the marked contradictions when compared on genetic grounds. The pathogen is having a dramatic lethal effect on the natural populations of Platanus orientalis kashmeriana in Northern India and solid containment measures should be imposed to restrict its spread throughout the natural range of this ecologically, economically and historically precious host.
Keywords: platanus orientalis kashmeriana, genetic change, dna barcoding, alternaria kashmeriana
Platanus orientalis L., the oriental plane perennial woody tree1 with important riparian species2 is widely spread in Crete and other southern islands. Its natural spread ranges from the southern Balkans, Crete, mountains of Turkey, Syria, Iran and Iraq, eastwardly to Kashmir valley, where it is familiarly known as the ‘chinar’. Chinar, Platanus orientalis kashmeriana, is the lone species of family Platanaceae found in India and growing throughout the valley of Kashmir. The family, Platanaceae consists of only a single living genus Platanus and is a native of the eastern Mediterranean portion from where it spread eastwards. It is a large deciduous tree, native to temperate regions (Huxley, 1992). Pharmacologically, the buds are used in folk medicine as antiseptic and antimicrobial medication of the urinary system.1 Plane has distinct advantages such as rapid growth rate, easy propagation, lush foliage and also strong air purification ability. Canker stain of plane tree, caused by the fungus Ceratocystis platani is a lethal disease that attacks Platanus sp3 was earlier reported from the United States and Europe.4–6 The disease results in staining of the xylem, distraction of osmotic movement, cankers, and finally death of the tree.4 The first report of the disease was observed in Greece in the 2003 on oriental plane in the southwestern Peloponnese,7 where C. platani has caused considerable mortality of the oriental plane in natural stands such as streams, rivers as well as in ornamental plantings in London plane.7 During the mycological explorations to the northern India (Kashmir), similar canker stain type symptoms were observed on beneath of the leaf surfaces of Platanus orientalis kashmeriana to what has been reported earlier in United States and Europe. In this study, the genetic variation was assessed in nuclear rDNA in order to recruit the pathogens classification purely relied on the molecular myco taxonomic characterization based on fungal ITS DNA sequencing and its phylogenetic interpretations. The current revolutions in molecular biology have provided advanced techniques in myco taxonomy. Molecular methods exploit the naturally occurring variations in the DNA. In eukaryotes, the ribosomal DNA (rDNA) found in the nuclear genome in general consists of tandem repeats of three RNA genes which undergoes transcription as a single unit and code for the 18S, 5.8S, and 28S subunits of the RNA, (Figure 1).8 The internal transcribed spacer (ITS) region of the nuclear ribosomal DNA is the unique DNA barcoding site for molecular identification of fungi. Analysis of DNA sequences from rRNA genes and the ITS region have been used in studies of phylogenetic relationships and evolution of a wide range of organisms. The gold standard for fungal ITS sequence based classification rely on the concept of phylogenetic recognition of species. This systematic approach is commonly used with conserved large or small subunit of rDNA across taxonomically diverse fungi.9,10 The inclusion of taxonomically sundry ITS sequences into a single alignment are hectic because of considerable amounts of sequence variation, normally only the highly conserved 5.8S region can be considered for phylogenetic interpretations. Specific ITS primers (ITS1 and ITS4) were used in PCR to amplify a portion of the 18S rRNA gene, the ITS1 and ITS2 regions. Variation within the 18S rDNA and ITS1 region was interpreted viz sequence analysis and by digesting the amplified rDNA to give allele specific restriction fragment patterns. Molecular characterization of ITS region of rDNA of causal organism revealed the emergence of a new fungal species, Alternaria kashmeriana sp.nov., which may have evolved from closely related taxa encountering the ‘0.5’ of genetic change encountered by different bootstrapping values (random sampling with replacement) under geographically different environment.10–15
Tissue sampling
Periodical mycological visits were carried out to the selected locations. The fungal strains isolated from infected samples of Platanus orientalis kashmeriana collected from the different selected sites viz. Anantnag (04 isolates), Baramulla (03 isolates), Kupwara (05 isolates) and Srinagar (07 isolates). The Symptoms appeared on lower leaf surfaces as light dark brown irregular canker spots (like rust), later becoming deep grayish brown on the upper surface. Mostly the infected trees were prematurely defoliated. It was also observed that the extent of infection changes parabolically with the fluctuating environmental conditions in different seasons in the prevailing area (Figure 2). The fungal cultures were grown and maintained in PDA medium and for liquid culture media all fungal cultures were sub cultured in PDA kept in controlled environment growth chambers under continuous florescent light for 9-11 days at temperature of 22- 28°C. Fast growing olivaceous black to grayish in color and are suede like to floccose colonies. Microscopically, acropetal branched chains (blastocatenate) of multi celled conidia (dictyoconidia) and are produced sympodially, (Figure 3). Obclavate, obpyriform conidia sometimes ovoid or ellipsoidal with a short conical beak. On the 16th day fungal cultures were examined for CFU using hemolytic meter and the spore suspension of 1.103 concentrations were inoculated in potato dextrose broth medium.15-20

Figure 2 Magnitude of Alternaria kashmeriana occurrence and extent of its infection changes parabolically with the fluctuating environmental conditions in different seasons in the prevailing area.

Figure 3 Shows 10 days old homogenous culture of Alternaria kashmeriana on PDA/6-Benzyl Amino Purine/Malt Extract Agar, incubated at 22- 28°C. Fungal cultures were examined for CFU using hemo c meter and the spore suspension of 1x103 concentrations.
DNA isolation
According to Molar, 50mg of dry mycelium mat was transferred in a sterile Eppendorf tube. It was crush with sterile scissor and made fine powder. 600μl of 60°C pre-warmed CTAB extraction buffer was added. The resulted mycelial powder was incubated in a water bath at 50°C for 3-4 hours with vortexing at the interval of 10 minutes. The resulting samples were centrifuged at 10000 rpm for 20 minutes at room temperature. The supernatant transferred to sterile Eppendorf tube. 600μl of Chloroform and isoamyl alcohol (24:1) were added and subjected to vortexing again for 5 minutes. 0.5μl 3 M of sodium acetate (pH 5.2) was added and mix gently and stored on ice for 15 minutes. The solution was again centrifuged at 16000 rpm at 4°C for 10 minutes. 600μl of chilled isopropanol was added to Eppendorf tube and stored at -50°C for 10 minutes. The supernatant decanted and 600μl 70% ethanol was added and solution was kept at room temperature. The overnight pellet was dried. 25μl of distilled water was added and kept at room temperatures for 2 hours followed by electrophoresis and nano drop spectrophotometry. The DNA samples were stored at -20°C.
PCR amplification and digestion of rDNA (ITS Gene Amplification)
ITS gene was amplified by selecting specific primer and optimizing PCR conditions. The primers (ITS1 and ITS4) showed highest amplification efficiency (Table 1). PCR reactions were performed using Kapabiosystems kit in 96-well plates. The reaction master mix constitute of 9.6 µl of 10% trehalose, 7.0µl H2O, 2.5 µl 10X PCR buffer ‘B’, 0.8 µl MgCl2, 2.0µl 2.5mM DNTP, 1.0 µl 10mM forward and reverse primers each and 0.1 µl taq polymerase (5 units/µl). This master mix is then distributed in each PCR tube 24µl of master mix in each PCR tube with 1µl DNA template having concentration of 30-100ng/µl. The PCR reaction thermal cycles includes an initial step of 3 min at 95°C and 35 cycles of 30 sec at 94°C, 40 sec at 52°C, and 1 min at 72°C, with a final extension at 72°C for 10 min. Amplicons were visualized on 1.5% agarose gel shown in (Figure 4).
Primers |
Sequence (5' - 3') |
ITS1 |
TCCGTAGGTGAACCTGCGG |
ITS4 |
TCCTCCGCTTATTGATATGC |
Table 1 Primers used for PCR amplification of ITS gene
Internal Transcribed Spacer (ITS) Sequencing
These PCR amplicons were processed for cleanup to remove unincorporated nucleotide and residual primers using Exo-SAP enzyme. These amplicons were forwarded for cycle sequencing reaction using BigDye® Terminator v.3.1 Cycle Sequencing Kit (Applied Biosystems, Inc.) with 16th fold dilution. For sequencing of ITS1&ITS4 primers were used for gene amplification were successfully used here. The thermal cycler conditions were an initial 5 step of 2 min at 96°C and 35 cycles of 30sec at 96°C, 15 sec at 55°C and 4 min at 60°C. The Cycle sequencing is followed by sequencing cleanup by ethanol precipitation method followed by dissolving template in Hi-Di formamide. These samples were bi directionally sequenced in ABI 3130 Genetic analyzer.
Nucleotide sequence accession numbers
The Gene Bank accession number for the sequence of the ITS region product using primers ITS1 and ITS4 in PCR amplification from Alternaria kashmeriana sp.nov. Strain 150b (NCBI unique submission Id BankIt1858984) were assigned NCBI GenBank accession number KT825857.
Phylogenetic analysis
The Phylogenetic tree has documented enough information about the inferred evolutionary relationships among the different allied Alternaria species. The ITS sequence was BLAST using data base to render the species similarities. The maximum likelihood analysis carried out by MOLE BLAST and statistical values of relatedness given by bootstrapping (random sampling with replacement) using another bioinformatics program The bottom line segment with the numeric number '0.5' reflects the length of branch which represents an amount of genetic change. The branch length units (nucleotide substitutions/site) are the number of genetic changes or substitutions divided by the sequence length, Figure 5. A branch length fluctuates proportionally to the nucleotide differences, and the numbers indicated on respective branches reflects the percentages of frequencies with which a given branch appeared in different 100 bootstrap replications.
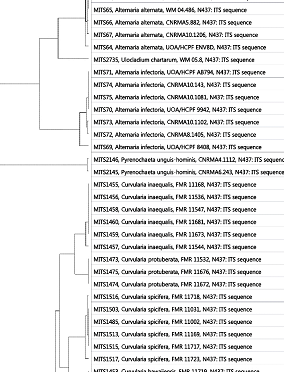

Figure 5 Maximum likelihood phylogenetic tree based on small subunit–internal transcribed spacer–large subunit (SSU-ITS-LSU) rDNA sequence variants of the Alternaria kashmeriana with the closely related sequence matches constructed by phylogeny.lirmm.fr. Branches receiving<70% bootstrap support were collapsed to polynomials, and long branches were shortened by 40%, which is indicated with two diagonal slashes. Bootstrap values are given for branches among but not within cultures. The scale bar indicates the number of substitutions per site with the number '0.5' shows the length of branch that represents an amount genetic change.
BLAST Score (E-Value summary)
The E-value or Expect value is a metric tool that has shown the significant alignment in a particular homology ITS sequence match. In the specific BLAST search results are designated an E-value which reflects how significant the BLAST hit is.
Statistically, the E- value is calculated as:
Where, K and λ are constants which purely depend on the used scoring matrix, while as m is the query length and n is the database length and S is the alignment score. The calculated E-values of different hits are given in Figure 6(a) & 6(b).
The next generation sequencing technologies can be expected to have a tremendous effect on fungal biodiversity and ecology research. In order to realize the full potential of these exciting techniques by accelerating biodiversity assessments, identification procedures of Alternaria species need to be adapted to the emerging demands of modern large-scale ecological studies. The ITS region has been used as the de facto standard ‘barcoding’ marker by mycologists for many years, and there is every reason to believe that it will remain at least one of the main fungal barcoding markers for many years to come. Further, the molecular myco taxonomic characterizations of Alternaria Kashmeriana sp. nov. revealed ample amount of genetic divergences probably due to the persistent extremely harsh environmental conditions in the surveyed area. Being deciduous nature of flora, the fungal growth may have encountered the extremes of harsh weather viz. prolonged chilly snow periods prevailing in the region which might have result in variations in fungus genotype as well as phenotype. Direct analysis/interpretations of DNA polymorphisms are a useful method to ascertain genetic variation in Alternaria species. Sequence analysis from ITS regions has made some progress towards isolation and better understanding the taxonomy of Alternaria kashmeriana sp.nov but further molecular analysis using more sensitive techniques to assess genetic variation have been implemented to determine the extent to which different taxa are present within this complex. In the present study ITS region of rDNA gene complex was used to evaluate genetic diversity/variation of geographically distributed Alternaria isolates. Sequence specific primers were designed at conserved genic regions of rDNA gene complex to amplify ITS regions. Phylogenetic analysis emphasized the evolutionary distance between different Alternaria species and related taxa. Alternaria kashmeriana sp.nov and Alternaria alternata are more closely related to each other than to Alternaria infectoria MITS71 isolates. It is sometimes difficult to differentiate the Alternaria kashmeriana sp.nov and from the rest known Alternaria species on the phenotypic bases but reflects the marked contradictions when compared on genetic grounds. Further the bootstrapping allowed to assign the measures of accuracy to sample estimates of Alternaria kashmeriana sp.nov with the rest of Alternaria species. The variance of 0.12655 of Alternaria kashmeriana sp.nov. From the closely related Alternaria isolates has been recorded with boot values of 0.2531, 0.07668, and 0.21472 (marked as SEQ00004, SEQ00002, SEQ00003 respectively (Figure 6b)). Therefore it is concluded that the causal organism of canker stain in India is Alternaria kashmeriana sp.nov which has been evolved with genetic variance/mutation of 0.5 in ITS region of rDNA from closely related species under geographically distinct region (The great Himalayan mountains).
The invasion or evolution and diversification of new strains or species under geographically distinct regions with different and extremely harsh environments are a lethal threat to disease prone regions which are comparatively less explored. The present study revealed the isolation of a new fungal pathogen, its identification and classification supported by both classical as well as molecular techniques. To our knowledge, this is the first report of canker stain disease of Platanus Orientalis Kashmeriana so far, caused by new fungal species. The disease is currently limited to the northern India where it was first observed. However, it may become epidemic due to its wide host range.
We thank Chief Conservator, Jammu & Kashmir for access to forest surveys, Central laboratory Instrumentation Centre (CLIC), Dr. Harisingh Gour Central University, Sagar, India and Director, Paul Hebert Centre, Aurangabad for DNA Barcoding and genetic analysis. This work was financially supported by the University Grants Commission (UGC), India, sponsored Central University fellowship.
The authors declare that no conflicts of interest exist for publication of this manuscript.

©2017 Dar, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.