MOJ
eISSN: 2374-6912


Research Article Volume 4 Issue 4
1Department of Microbiology, AJA University of Medical Sciences, Iran
2Department of Bacteriology, Tarbiat Modares University, Iran
Correspondence: Majid Eslami, Department of Microbiology, AJA University of Medical Sciences, Iran
Received: August 08, 2017 | Published: August 11, 2017
Citation: Ghasemian A, Nojoomi F, Eslami M. The Rep-PCR typing of TEM type ESBL producing clinical isolates of klebsiella pneumonia. MOJ Cell Sci Rep. 2017;4(4):98-102. DOI: 10.15406/mojcsr.2017.04.00092
Objective: The aims of this study were to detect and fingerprint TEM-producing clinical isolates of K pneumonia.
Materials and methods: In this cross-sectional study from July 2013-May 2015, a total of 200K. pneumonia isolates were collected from various origins (all from hospital acquired infections) in Tehran, Iran. The antibiotic susceptibility and MIC of isolates were performed according to the CLSI guidelines. The combined disk method was employed to determine ESBL producer isolates. PCR was done for detection of the gene encoding TEM type enzyme. The rep-PCR technique was applied for the fingerprinting of K. pneumonia isolates.
Results: The age range of patients was 1-69 years (mean= 41.33, including 93 (46.5%) males and 107 (53.5%) females). Ninety (45%) isolates exhibited MIC≥2 to ceftazidime and cefotaxime, respectively, and 87 (43.5%) demonstrated ESBL production. The highest and lowest resistance was against erythromycin and imipenem (93.5% and 0%, respectively). The prevalence of bla TEM gene among ESBL producers was 87.3% (n=76). The rep-PCR typing of isolates showed diversity but a 90% similarity among TEM producing isolates, suggesting polyclonal spread of TEM type producing isolates.
Conclusion: The findings of the current study concluded the emergence and spread of K. pneumonia isolates producing TEM type enzymes with a genetically diverse background in Tehran.
Keywords: klebsiella pneumonia, tem type esbls, genotyping
Extended-spectrum Beta-lactamase (ESBL) producing K. pneumonia has been considered among six drug-resistant microbes to which new therapies are urgently needed.1 Increasing resistance in K. pneumoniae to first-line antibiotics have made therapeutic options for urinary tract (UTIs) and other infections challenging.2 The reports of ESBLs from K. pneumonia have been sharply increased since the years 2009-2010 until today.3,4 TEM- and SHV-ESBLs, with over 100 mutations being reported, are major cause of hospital-acquired infections, particularly in the Intensive Care Unit (ICU). The blaTEM genes are parts of the Tn3 transposons elements, namely, Tn3 (blaTEM-1a), Tn1/Tn801 (blaTEM-2) and Tn2 (blaTEM-1b), which are carried by highly transmissible plasmids.5,6 Although TEM-1 was considered as the most frequent gene among ESBL producers in 1980s and early 1990, today there are reports showing SHV-1 as the most prevalent ESBL gene in many parts of the world.7 It has been supposed that the naturally occurring TEM-type ESBLs are the outcome of fluctuating selective pressure due to several beta-lactams within a given ward or hospital rather than selection with a single agent.8 Repetitive estrogenic Palindromic elements (REP) are short sequences mostly described in gram-negative species.9 The prevalence of K. pneumonia TEM type ESBLs among hospitalized patients in Iran have been reported as following: 32% (n=18) from Tehran in 2007,10 35.29% (n=18) from Tehran in 2008,11 54% (n=48) from Tehran in 2010,12 8% (n=7) from Tehran in 2010,13 15.38% (n=4) from Ahvaz in 2013, 30.7% (n = 32) from Tehran in 201014 and 39% (n=18) from Shiraz in 2013.15 Several studies have shown that rep-PCR has the ability to fingerprint strains of Escherichia coli and K. pneumonia and other gram-negative species.16-19 There has been excellent correlation between MLST and automated rep-PCR in K. pneumonia fingerprinting.20 The aims of this study were to detect and fingerprint TEM-producing K. pneumonia isolates from hospitalized patients in Tehran.
Bacterial strains and cultures
In this cross-sectional study from July 2013- May 2015, a total of 200 K. pneumonia isolates were collected from hospitalized patients with age range of 1-69 years (mean=41.33, including 93 (46.5%) males and 107 (53.5%) females) in Tehran, Iran during 2013-2015. The isolates were identified by biochemical tests and subsequently were stored in Trypticase soy broth containing 30% glycerol at -20ºC.
Antibiotic susceptibility testing The antibiotic susceptibility testing was conducted with disk diffusion method (Kirby-Bauer) according to CLSI recommendation using 14 antibiotics, including FOX (30µg), CAZ (30µg), CTX (30µg), CPM (50µg), ATM (50µg), ERY (15µg), GM (10µg), TE (30µg), SXT (25µg), AX (30µg), AM (25µg), IPM (10µg), AN (30µg) and CP (30µg) (from MAST, UK), determined. E.coli ATCC 25922 was used to control antibiogram and combined disk.21
Phenotypic ESBL production
ESBL production was assessed by using combined disk method, in which cefotaxime (30µg) and ceftazidime (30µg) with and without the Clavulanic acid were used. After incubation for 24 hours in 37ºC ESBL producing by increasing the size of inhibition zone diameter ≥5mm to Cefotaxime and Ceftazidime in the combination of each of them with Clavulanic acid were an indicator of ESBL production.22
Minimum inhibitory concentration (MIC)
The agar dilution MIC method with and without Clavulanic acid inhibitor to Cefotaxime and Ceftazidime in concentrations serial 0.5 to 512 µg/ml was used. E. coli ATCC25922 was used to control bacteria. MIC in the presence of 4 µg/ml of Clavulanic acid inhibitor was used in all plates.
DNA extraction
K. pneumonia isolates were cultured on trypticase soy agar for 24 h at 37ºC. DNA from each strain was extracted using the Cinagen kit according to the manufacturer’s instruction. DNA yield was determined based on absorbance at 260nm using a spectrophotometer and stored at -20°C.
PCR detection of blaTEM gene
TEM (F) 5'-ATG AGT ATT CAA CAT TTC CG-3' and TEM (R) 5'-CCA ATG CTT AAT CAG TGA GG-3' primers were used for the amplification of genes coding for TEM type β-lactamase. Amplification was performed in a final volume of 25µl containing 0.5 U of Taq DNA polymerase, 2.5 µl of 10 × buffers, 2.5 µl dNTPs, 2 µl primers (12.5pmol each), and 2.5µl DNA template. Reactions were run with an eppendorfs PCR system under the following conditions: 4 min at 94 ºC, followed by 35 cycles of 1 min at 94ºC, 1 min at 55ºC, and 1 min at 72 ºC, followed by a final extension of 10 min at 72ºC. The PCR products were electrophoreses on 1% agarose gel (sigma), stained with ethidium bromide, and photographed under UV light.
Rep-PCR typing
DNA was subjected to PCR utilizing the primers REP 1R-I (5'- IIIICGICGICATCIGGC -3') and REP 2-I (5'- ICGICTTATCIGGCCTAC -3') as described by Versalovic et al. [23]. Amplification reactions were performed in a final volume of 25 µl containing 45pmol (1.8µmol.L-1) and 36pmol (1.44 µmol.L-1) of each of the REP primers, 0.200mmol.L-1 of each deoxynucleoside triphosphate, 10mmol.L-1 Tris-HCL (PH8.3), 1.5 mmol.L-1 MgCl2 and 0.35 U of Taq DNA polymerase. Amplification was performed in a PCR thermal cycler, using the following temperature profile: 95ºC for 6 min, 30 cycles at 94ºC for 1min, at 40ºC (REP-PCR) for 1 min and at 65ºC for 8 min, followed by a final extension of 16 min at 65ºC. PCR products (10 µl) were separated on 2% agarose gel (sigma) containing 1 × TBE buffer (0.1 mol.L-1 Tris-HCL, 0.1mol.L-1 boric acid, 0.002 mol.L-1 EDTA, PH= 8) at 4 V.CM-1. After rep-PCR, gel images were transferred to Phoretix 1D software. Rolling ball method was used to remove the background. Identification of the bands was performed manually. The standard strain (E.coli ATCC 25922) was used to determine the lines Rf (standardization bands between different samples). Bands matching with error rate of 0.02 were done. Code 0 (no band) and 1(band) from the program were transferred to NTSYSpc software and finally drew a dendrogram of Dice similarity coefficient with the Complete Linkage algorithm.
Statistical data analysis
Data was analyzed by employment of Graph Pad Prism version 6.1, which t-test and F-test were applied where needed. The confidence interval equal to 95% (CI=95%) was considered and P value˂0.05 was significant.
Susceptibility pattern
Percentage of isolates resistant to antibiotics in this survey in the disk diffusion method was as following: FOX 46.5%, CAZ 45%, CTX 72%, CPM 4.5%, ATM 39.5%, ERY 93.5%, GM 36.5%, TE 75%, SXT 57%, AX 85%, AM 91%, AN 15.5%, IPM 0.5%, and CP 39%. The majority (n=76) were resistant to Ampicillin, whereas the lowest resistance was shown to imipenem.
CAZ and CTX MIC of isolates
Serial concentrations of 0.5 to ≥512 µg/ml of Ceftazidime were prepared and tested. Strains resistant to Ceftazidime MIC for the combination were Clavulanic acid. In this experiment, 90 isolates of K. pneumonia and MICCAZ ≥16 were considered resistant. The use of Clavulanic acid inhibitors this number was reduced to 18 isolated (Tables 1 & 2). Cefotaxime MIC levels among the isolates have been shown in Table 2. Cefotaxime MIC for the resistant strains was done with and without Clavulanic acid. In this test, 145 isolates with MIC CTX ≥ 4 were detected and 65 isolates were Clavulanic acid susceptible.
µg/ml |
≤ 4 |
8 |
16-32 |
64-128 |
256-512 |
> 512 |
CAZ |
83 |
27 |
66 |
13 |
1 |
10 |
CAZ + CA |
149 |
33 |
8 |
4 |
2 |
4 |
Table 1 Ceftazidime MIC in the presence and without the presence of Clavulanic acid for K. pneumonia isolates
µg/ml |
1≥ |
2 |
8-Apr |
16-32 |
64-128 |
256-512 |
≥ 512 |
CTX |
49 |
6 |
48 |
21 |
34 |
21 |
21 |
CTX + CA |
129 |
6 |
58 |
0 |
1 |
0 |
6 |
Table 2 Cefotaxime MIC in the presence and without the presence of Clavulanic acid for E. coli isolates
PCR test for the blaTEM gene
Eighty-four (54.1%) of 155 isolates were examined for TEM enzyme gene. The highest percentage of this gene was detected in urine samples and the lowest percentage was in blood. The distribution of TEM type enzyme in various clinical origins has been exhibited in Figure 1. Percentage Of antibiotic resistance in isolates with TEM positive has been shown in Figure 2. PCR products for TEM-lactamase gene has been shown in Figure 3. Rep-PCR typing of isolates containing TEM gene. Dendrogram plotted using the Complete Linkage algorithm and Dice similarity coefficient Phoretix 1D software and NTSYSpc (version 2.02) for the TEM gene showed that K. pneumonia placed in 6 clusters (A-F, Figure 4). Most of urinary isolates placed in cluster A showing a genetic relation among them. Moreover, all were susceptible to imipenem and the majority to amikacin. These isolates showed MICCAZ and MICCTX equal or lower than 16µg/ml. The homology of more than 90% was observed among the majority of isolates and two of them showed 100% homology and were isolated from wound infection. The phenotypic similarity of these two isolates was also more than 90%. Isolates within each of other clusters also showed similarities like these descriptions (Figure 5). A similarity of 90% was observed among 50-80% of isolates. The relation of rep-PCR clusters to drug resistance and infection sites has been depicted in Table 3.
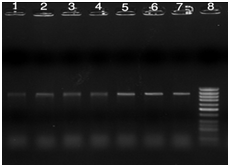
Figure 3 PCR products of TEM gene with 850bp size, Lanes 1-6: positive clinical samples, Lane 7: positive control, and lane 8: 100bp DNA Ladder.
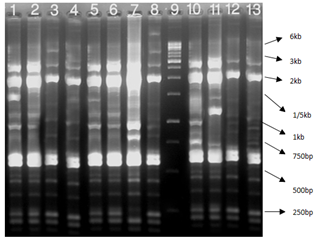
Figure 4 Rep-PCR reactions for genotyping of TEM positive K. pneumonia: Lane 1 positive control E.coli ATCC25922, Lane 2-11 clinical samples, and Lane 12 1kb DNA Ladder.
Variable |
Cluster A (n=21) |
Cluster B (n=8) |
Cluster C (n=7) |
Cluster D (n=19) |
Cluster E (n=6) |
Cluster F (n=19) |
MDR strains (n=30) |
9 |
3 |
1 |
9 |
1 |
8 |
Stool (n=11) |
3 |
1 |
0 |
5 |
0 |
2 |
Urine (n=24) |
9 |
1 |
1 |
4 |
2 |
7 |
Blood (n=6) |
4 |
1 |
0 |
1 |
0 |
0 |
Tissue (n=13) |
4 |
0 |
1 |
2 |
2 |
4 |
Wound (n=17) |
1 |
2 |
3 |
4 |
2 |
5 |
Secretions (n=9) |
0 |
3 |
2 |
3 |
0 |
1 |
Table 3 The relation of rep-PCR clusters to drug resistance and infection sites
MDR: Multiple Drug Resistant; ICU: Intensive Care Unit; ID: Infectious Disease
In this study, among hospitalized patients the majority of isolates were resistant to ampicillin, erythromycin, co-amoxiclav, but more than 90% were susceptible to imipenem. Other studies have also shown a high resistance to these antimicrobials worldwide.24,25 However, carbapenem-resistant K. pneumonia has been frequently isolated and developed.26,27 Resistance to amikacin was observed among 15.5% of isolates. We observed a correlation between antibiotic susceptibility and each rep cluster. A study by Duin28 showed a relation between rep cluster type A and KPC producing K. pneumonia, in which most of positive isolates placed in cluster A, although two isolates showed different susceptibilities to amino glycosides and tige cyclones. In this study, 76/200 (87.3%) of isolates amplified the blaTEM gene. The rep-PCR pattern depicted six clusters (A-F), which mostly classified in clusters A, D and F, whereas a minority classified in clusters B and C. The genetic background of isolates was different in spite of 90% similarity of 50-80% of them and thus it was suggested a polyclonal spread of TEM type producing isolates in a study from Portugal, CTX-M and TEM type producing K. pneumonia isolates were genetically diverse.29 In Mensah’ study in Ghana, 48.1% of K. pneumonia and E. coli isolates were TEM positive, but the genetic relationship of isolates has not been investigated.30 To the best of our knowledge no previous studies have determined genetic relationship among TEM type producing K. pneumonia isolates. In a study by Shakibaei from Kerman, only 2.5% of K. pneumonia isolates amplified TEM enzyme, but strain typing has not been elucidated.31 Another study by Mansury32 from Shiraz exhibited that 16% of K. pneumonia amplified TEM, but no typing method was performed. In a study in Tehran, among 43K. Pneumonia (14 ESBL positive and 29 ESBL negative) which adopted for RAPD-PCR typing, 46.5% belonged to a single profile (genotype 1), of which, the majority (62.1%) were non-ESBL producers.33
We observed that TEM type isolates from urine and blood specimens were mostly classified in cluster A (13/30), and those from stool, wound and secretions were classified in cluster D. moreover, 16 isolates from urine (n=7), tissue (n=4) and wound (n=5) were classified in cluster F (16/54). A study by Kathrin have exhibited that there is a possibility of relation between site of infections and genetic background, drug resistance or pathogenesis of Klebsiella spp.34 We appreciated that MDR K. pneumonia (isolates with resistance to beta lactams, quinolones and amino glycosides, n=30) distributed in 6 clusters in a diverse pattern. The limitations of this study were lack of typing with more accurate or reliable methods such as pulsed field gel electrophoresis (PFGE) and multi-locus sequence typing (MLST), no data of hospital settings for some reasons, low number of TEM type producing K. pneumonia isolates and a narrow area of study.
The findings of the current study concluded the emergence and spread of K. pneumonia isolates producing TEM type enzymes with a genetically diverse background in Tehran.
This study was approved by ethical committee of Bandar Abbas University of Medical Sciences, (Grant No. 1649547/6743, 2015).
None
The author declares no conflict of interest.

©2017 Ghasemian, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.