MOJ
eISSN: 2374-6912


Mini Review Volume 4 Issue 3
Epi Gen Biotechnology Consultancy, Turkey
Correspondence: Burcu Biterge, Epi Gen Biotechnology Consultancy, Turkey
Received: April 29, 2017 | Published: June 12, 2017
Citation: Sut I, Biterge B. Acid-labile histone phosphorylations. MOJ Cell Sci Rep. 2017;4(3):76-77. DOI: 10.15406/mojcsr.2017.04.00088
Eukaryotic DNA is packaged into chromatin, folded and compacted. The building blocks of mammalian chromatin are nucleosomes, which consist of octomers of histone proteins that can be post-translationally modified (PTM). Protein phosphorylation is one of the most abundant PTMs. Histones are phosphorylated mainly on serine, threonine, tyrosine as well as other sites such as arginine, histidine and lysine that are not so well-known. Both types of protein phosphorylations occur in vivo at similar levels; however, phosphoarginine, phosphohistidine and phospholysine are thermodynamically unstable and therefore much less studied.
Keywords:chromatin, histone, post-translational modification, PTM, phosphorylation, acid-labile
PTM, posttranslational modification; ATP, adenosine triphosphate; P-N, phosphoramidate bond; AEBP1, adipocyte enhancer binding protein 1; HHK, histone h4 histidine kinase
All eukaryotic organisms face the challenge of fitting their DNA into the nucleus. To overcome this problem, genomic DNA is compacted into a macromolecular structure called chromatin in a dynamic manner so that it can still be accessible to carry out cellular functions such as replication, transcription and DNA repair. The first step of compaction is achieved by packaging the naked DNA into nucleosomes, which are the repeating unit of chromatin, by wrapping 147 base pairs of DNA around the histone octamer containing two copies of each core histone H2A, H2B, H3 and H4.1 Histones are small, basic proteins that are composed of a globular domain and unstructured N- or C-terminal tails that are subjected to several covalent modifications such as methylation, acetylation, and phosphorylation.2 Phosphate groups are added to and removed from the target histone residue by histone kinases and phosphatases respectively. The transfer of a phosphate group from ATP to the hydroxyl group of the amino acid side chain introduces a negative charge, which can affect electrostatic interactions within chromatin.3 So far, most attention had been focused on Serine, Threonine and Tyrosine residue phosphorylation. These modifications are known to be stable under acidic pH and labile at high pH owing to the stability of the P-O bond generated by the addition of a phosphate group.4 However, there is another type of phosphorylation, which is widely neglected yet, but equally important. This second class of phosphorylated amino acids comprises Arginine, Lysine and Histidine residues (Figure 1). Phosphorylation of these amino acids results in the formation of a P-N (phosphoramidate) bond, which is, unlike the P-O bond, acid-labile and base stable.5 Although these phosphorylations occur naturally in comparable levels, they have been usually missed since many techniques classically used to study protein phosphorylation involve acidic conditions.6 Furthermore, it has been relatively more challenging to study the functions of acid labile histone phosphorylations since the phosphate groups are quickly hydrolyzed when peptides containing these modifications are used as immunogens to raise site-specific antibodies, which makes it harder to study their physiological roles. For this reason, new methods to synthesize phosphohistidine and phosphoarginine peptides with improved thermodynamic stability have been developed.6,7 This review focuses on 2 major types of acid-labile histone phosphorylations, occurring on arginine and histidine.
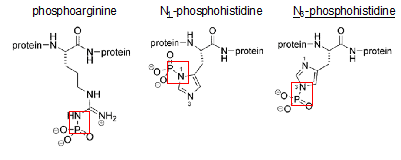
Figure 1 Phosphorylation of Arginine, Histidine and Lysine. Red squares mark the acid labile P-N bond set upon phosphorylation.
Histone arginine phosphorylation
First reports on protein arginine kinases date back to the late eighties. A cAMP-independent protein kinase was identified to phosphorylate an 11-kD chromosomal protein at an arginine residue in 1987.8 Then, a calmodulin activated histone H3 kinase has been partially purified from calf thymus and they were later on able to show that the histone H3 is phosphorylated at four different sites; arginines 2 (N-terminal), 128, 129 and 131 (C-terminal) by a partially purified mouse leukemia cell line kinase.9,10 In a follow up study by the same authors, a histone H3 kinase from both quiescent and dividing rat endothelial cells was identified as an 85-kD protein, which was further proven to be a protein called Adipocyte Enhancer Binding Protein 1 (AEBP1)11 and unpublished data). AEBP1 was originally characterized as a transcriptional repressor that binds to the adipocyte enhancer-1 site of the aP2 promoter, which encodes an important marker of adipocyte differentiation.12,13 One of the most interesting features of this protein is that it has a carboxypeptidase domain, which is stimulated by DNA binding and was also shown to be important for the repressor activity.14 Although the AEBP1 protein has been widely studied as a transcriptional repressor and for its carboxypeptidase activity, its potential role as an arginine kinase has not been evaluated.
Histone histidine phosphorylation
Histidine kinases have previously been shown to have important roles in signal transduction both in prokaryotes and lower eukaryotes.15 These so called “two-component histidine kinases” are usually composed of a membrane bound receptor-like protein, which has the kinase domain and will be activated upon stimulation and a response regulator protein such as a transcription factor, which will be receiving the phosphoryl group from the phosphohistidine of the first component.16 The presence of a histone H4 histidine kinase (HHK) has been known since the seventies17 and recently Besant et al.18 published that the histone H4 can be phosphorylated both at histidine 18 and 75.18 In spite of the fact that they were able to link the HHK activity to proliferating and also tumorigenic cells, the biological role of the phosphohistidine as a histone modification remains unknown. Furthermore, the HHK activity was found to be elevated in hepatocellular carcinoma cells in comparison to the surrounding normal tissue, suggesting a role for phosphohistidine as an oncogenic marker.19
The eukaryotic phospho-proteome is composed of proteins phosphorylated at serine, threonine and tyrosine (acid-stable) as well as lysine, arginine and histidine (acid-labile) residues. Both classes of phosphorylations are stable under physiological conditions and occur in vivo at comparable levels. However, the difficulty of studying acid-labile histone phosphorylations prevented the role of these modifications from being completely understood. Therefore, future studies will shed more light on the chromatin regulation via acid-labile phosphorylations and contribute to our current knowledge of epigenetic mechanisms.
None.
The author declares no conflict of interest.

©2017 Sut, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.