MOJ
eISSN: 2374-6912


Research Article Volume 4 Issue 1
Centre of Systems Biology and Bioinformatics, Punjab University, India
Correspondence: Ashok Kumar, Centre of Systems Biology and Bioinformatics, Panjab University, India
Received: March 18, 2016 | Published: January 26, 2017
Citation: Sood S, Kumar A. Identification of fusarium moniliformin pg inhibitor using comparitive bioinformatics studies of PGIP of Pharsalus vulgaris and its neighbours of LRR superfamily. MOJ Cell Sci Rep. 2017;4(1):5-11. DOI: 10.15406/mojcsr.2017.04.00075
Invasion of the Phytopathogenic organisms such as fungi, bacteria, virus and nematodes which disrupt the cell wall of the plant by secreting their pectin degrading PolyGalacturonase (PG) enzyme is evaded by the interaction of plant PolyGalacturonase-Inhibiting Proteins (PGIPs). PGIP from Pharsalus vulgaris is reported to blocks the attack of many pathogens, most importantly Fusarium moniliforme. The two PGIP isoforms, PGIP1 and PGIP2 sharing 99% sequence identity show differential activity towards FmPG. PGIP2 blocks FmPG while PGIP1 is unable to do so, while both these proteins belong to the same multigame family. PG-inhibitory activity is also shown by PGIPs of other plant species. Identification of sequence homolog’s of PvPGIP2, sequence similarity of at least 50%, through BLAST followed by MSA and nearest neighbour analysis suggests Glycine max (Soybean) and Pisum Sativum (Pea) to be close relatives of Pharsalus vulgaris (Kidney Bean). As already known GmPGIP3 effectively evades infection caused by Fusarium moniliforme blocking its PG enzyme while in Pea the efficacy of PGIP1 against FmPG is yet to be explored. Pairwise sequence comparisons and structure modeling of PvPGIP1 and PsPGIP1 is followed by prediction of putative binding sites of the fungal enzyme FmPG by CASTp. A comparative analysis is then drawn by docking the three PGIP proteins from Kidney Bean (PvPGIP2 and PvPGIP1) and Pea (PsPGIP1) respectively with FmPG using GRAMM-X server. It brings into light that PsPGIP1-FmPG shows same mode of binding as PvPGIP2-FmPG complex despite of being less similar at sequence level. Further on, the residues involved in hydrophobic and ionic interaction are calculated for all the three docked complexes involving FmPG and PGIP proteins from Pharsalus vulgaris (PvPGIP1 and PvPGIP2) and Pisum Sativum (PsPGIP1) by PIC. The docking results reveal that PsPGIP1 from Pisum sativum blocks the active cleft of the fungal enzyme FmPG inhibiting the enzyme in similar way as shown by PGIP2 from Pharsalus vulgaris also the outcomes obtained from the interaction analysis suggest that since same residues of the enzyme FmPG are involved in hydrophobic and ionic interactions in the FmPG-PvPGIP2 complex and FmPG-PsPGIP1 thus PsPGIP1 from Pisum Sativum is able to block Polygalacturonase enzyme (FmPG) from Fusarium moniliformin.
Keywords:PGIP, FmPG; LRR, pharsalus vulgaris, fusarium moniliformin
PGIPs, polygalacturonase-inhibiting proteins; PG, polygalacturonase; PV, pharsalus vulgaris; PAMPs, pathogen-associated molecular patterns; FM, fusarium moniliformin
Plants are exploited as a source of food and shelter by a wide range of parasites, including viruses, bacteria, fungi, nematodes, insects and even other plants. Plants lack a circulating adaptive immune system to protect themselves against pathogens hence they have evolved other mechanisms of antimicrobial defence which are either constitutive or inducible.1 Pathogen recognition by plants maybe in the non-host and host specific resistance manner. In host defence, plant disease resistance (R) genes encode proteins that bind specific pathogen-derived a virulence (Avr) protein and in non-host resistance, specific pathogen or plant cell wall derived exogenous or endogenous elicitors are recognized. Pathogen-associated molecular patterns (PAMPs) are conserved microbial molecules, such as lipopolysaccharides, peptidoglycans and flagellin of bacteria, mannans of yeast, chitin and ergosterol of fungi; they perceive pathogens and activate innate immune responses in plants. The defence resistance (R) genes, located either on the plasma membrane or in the cytosol.2–4 These R genes are found to encode five classes of proteins with certain common motifs, namely intracellular protein kinases; receptor-like protein kinases with an extracellular leucine-rich repeat (LRR) domain; intracellular LRR proteins with a nucleotide binding site (NBS) and a leucine zipper (LZ) motif; intracellular NBS-LRR proteins with a region with similarity to the Toll and interleukin-1 receptor (TIR) proteins; and LRR proteins that encode membrane bound extracellular proteins. Plant proteins with extracellular leucine-rich repeats are the players of innate immunity in plants and are known as Polygalacturonase-Inhibiting Proteins (PGIPs). These are found on cell surface of plant cells and inhibit the enzyme Polygalacturonase (PG). The enzyme pectinolytic in nature is expressed and secreted by phytopathagenic fungi at the early stages of plant infection; degrade homogalacturonan, the main component of cell wall pectin, a major component of the primary walls of dicotyledonous and non-graminaceous monocotyledonous cells, and cause cell wall degradation and tissue maceration.5 PGs are known to be secreted by phytopathagenic fungi, bacteria and nematodes.
Plants secrete proteins that specifically bind to PGs and modify their enzyme action6 and are known as PolyGalacturonase-inhibiting proteins (PGIP). Polygalacturonase-inhibiting proteins (PGIPs) are extracellular leucine-rich repeat (eLRR) proteins that recognize and inhibit fungal PolyGalacturonase (PGs) and are typically effective against fungal PGs.7,8 The first gene encoding a PGIP was cloned from Pharsalus vulgaris.9 Pgip genes cloned from many plant species usually exist as gene families. At least two related genes are present in raspberry, tomato, pear, and apple.10 In the genome of P. vulgaris, a family of at least 5 genes is clustered on chromosome 10,11 as confirmed by the isolation of two overlapping BAC clones. Five different genes have been characterized to date namely, Pvpgip1, Pvpgip2.1, Pvpgip2.2, Pvpgip3 and Pvpgip4. The genes Pvpgip3 and Pvpgip4 are closely related to each other (93.4% identity), but share only 79% identity with Pvpgip1/Pvpgip2. The mature PGIP is characterized by the presence of 10 repeats, each derived from modifications of a 24- amino acid leucine-rich peptide. The leucine-rich repeat (LRR) element matches the extra cytoplasmic consensus GxIPxxLxxLxxLxxLxLxxNxLx,12 also found in several R genes, which participate in gene-for-gene resistance.13 LRR is a versatile structural motif responsible for many protein–protein interactions and involved in many different cell functions such as receptor dimerization, domain repulsion, regulation of adhesion and binding events.14 In plants, LRR proteins play a relevant role in both development and defence. The crystal structure of the only PGIP (PGIP2 from Pharsalus vulgaris) (Figure 1) as well as the first LRR protein belonging to the plant-specific subfamily reveals a typical curved and elongated shape. The central eLRR domain, folded in a right-handed super helix, consists of a set of ten tandemly repeating units made up of 24 residues and is characterized by these structural elements:
The concave surface is known to bear residues necessary for binding and recognition specificity in this class of protein. All known PGIPs are glycoproteins of molecular mass around 40KDa, and comprise 10 modules characterized by the consensus sequence for extracytoplasmic leucine-rich repeats (eLRRs), GxIPxxLGxLxxLxxLxLxxNxLT/S. Within each LRR the sequence xxLxLxx is predicted to form a β-strand/α-turn structure, in which the x residues are solvent-exposed and involved in the interaction with ligands.
PGIPs are specific inhibitors; they do not inhibit other cell-wall degrading enzymes, and the per cent inhibition of different fungal PGs differs thus PGIPs can discriminate between PGs and PGIPs from different plant sources differ in their inhibitory activities, but PGIPs from a single plant source also inhibit PGs from different fungi or different PGs from the same fungus, although with different strength. Variation in PG-PGIP interaction seems to reflect co-adaptations between fungal PGs and plant PGIPs leading to specialization.15 Analyses of PGIP nucleotide sequences cloned from different plants indicates that these genes show substantial homology, but their deduced amino acid sequences are sufficiently different to explain the functional variability in different plants. The amino acid sequence is divided into a set of 10.5 leucine-rich tandemly repeated units, each derived from modifications of a 24-amino acid peptide similar to that of the extracellular domain of a cloned Arabidopsis LRR like protein kinase (RLK 5), involved in protein-protein interactions.16 Detailed characterization of two bean PGIPs, PGIP-1 and PGIP-2, showed their effectiveness against Aspergillus Niger PG, whereas PGIP-2 was only effective against F. moniliforme PG. Such differences were mainly due to a small number of amino acid substitutions described a short peptide, corresponding to residues 181-244, located in the active cleft of endoPG from F. moniliforme responsible for interaction with bean PGIP. The complete Pspgip1 coding sequence is very similar to PGIPs described from several other plant species and is a 312bp protein.17 The cyst nematode Heterodera goettingiana is a host-specific pathogen of pea (Pisum sativum). After invading the plant roots, the nematode migrates intracellularly to the vascular cylinder where it selects a cell which becomes its initial feeding site. The cyst nematode penetrates the cell wall in a similar way as Phytopathogenic fungi or bacteria by degradation of the main cell wall components, including pectin and cellulose by secreting Polygalacturonases enzymes. A Pea PGIP (Pspgip1) gene is upregulated during nematode infection. PsPGIP1 contains 10 LRRs whose consensus sequence matches the extracytoplasmic LRR consensus sequence of resistance genes. The deduced protein also contains eight cysteine (Cyst) residues at conserved positions: four within the N-terminus, one in the 10th LRR and three within the C-terminus. These have been shown to form four disulphide bridges (Cys3-Cys33, Cys34-Cys43, Cys281-Cys303, and Cys305-Cys312) which are important for the maintenance of secondary structures in PGIP. The predicted mature protein sequence of the Pea PGIP1 showed a high degree of similarity (83%) with the sequence of P. vulgaris PGIP2 (PvPGIP2).
PGIPs are proteins which belong to the Leucine Rich Repeat (LRR) superfamily and play an important role in the plant immune responses. PGIPs evade the attack caused by fungal, bacterial, viral and other pathogens which infect the plant by penetrating through its cell wall. PGIP2 from Kidney Bean is highly characterized and crystallized protein which blocks the attack of many pathogens most importantly Fusarium moniliforme. The interaction of plant protein PGIP with FmPG is due to the presence of tandem repeats of 20-30 amino acid stretch of extracytoplasmic type with consensus sequence that bear a conserved part LxxLxLxxNxL or LxxLxLxxCxxL which form the Leucine Rich Repeat. The family members of Leucine-Rich Repeat (LRR) superfamily including Pharsalus vulgaris where the two PGIP isoforms, PGIP1 and PGIP2 sharing 99% sequence identity show differential activity towards FmPG. PGIP2 blocks FmPG while PGIP1 is unable to do so. PG-inhibitory activity is also shown by PGIPs of other plant species. In present study, we have performed database search of plant specific species. Using BLAST searches we have retrieved 28 plant species showing homology with PvPGIP protein sequence. Further, we have specifically aligned LRR domain regions of several PGIP proteins from various plant species. In our multiple sequence alignment result we have observed that Pisum sativum (Pea) was sharing a close relation with Pharsalus vulgaris as they belong to the similar leguminous group. Sequence identity with both the PvPGIP isoforms was high, hence we hypothesized whether PsPGIP1 be able to block the fungal enzyme FmPG or not?
The LRR region of Pharsalus vulgaris (PvPGIP1) and Pisum sativum (PsPGIP1) were further analyzed with neighbour joined tree method to establish sequence relationship for repeated regions of LRR domain. PGIP protein for Pea was modelled with homology based protein structure prediction server SWISS-MODEL. The fungal enzyme FmPG was examined for the presence of binding pocket amino acid residues and comparatively analyzed for the direct interaction with PGIP protein from Kidney Bean and Pea using protein-protein docking. Protein-protein docking studies were carried out using GRAMM-X server, a public web server for protein-protein docking. The PsPGIP1 interacts with FmPG and to block the active site of the enzyme to restrict its enzymatic activity. The interacting amino acid residues were identified by KFC server between the FmPG-PvPGIP1 complex, FmPG-PvPGIP2 complex and FmPG-PsPGIP1 complex. From experimental studies it was reported that it is not just the sequence that determines the activity of proteins but the hydrophobic and ionic interactions contribute significantly. The interactions are by virtue of the protein structure or in other words we can say that due to the presence of the LRR structural fold which gives rise to these interactions. Thus, the hydrophobic and ionic interactions were calculated using PIC: Protein Interaction Calculator and the comparative analysis of the interacting residues and interactions between the three complexes help us to decipher PsPGIP1 was able to block FmPG. In conclusion we have identified the PGIP1 protein from Pea successfully blocks the PG from F. moniliformin.
Retrieval of information
The crystallized structure and FASTA sequence of Pharsalusvulgaris PvPGIP2 (PDB ID: 1OGQ) and Endo PolyGalacturonase enzyme Fusarium moniliforme FmPG (1HG8) (Figure 2) was retrieved from PDB.
PGIP sequence analysis: PvPGIP2 (Pharsalus vulgaris) inhibits the fungal enzyme FmPG (Fusarium moniliforme) by blocking its active cleft. Although another protein belonging to the same multigene family namely PvPGIP1 sharing 99% sequence similarity with PvPGIP2 is unable to block FmPG and fails to prevent pathogenic infection as reported from previous studies.18 PGIPs from different or same plant differ in their inhibitory activity towards the same PG. UniProt BLAST search carried out in order to locate sequence homologs from various plant species using ungapped alignment so as to include LRR motif during alignment. Further, the selected homologs were subjected to MSA by CLUSTALW19 yielding a guide tree based on NJ approach. The phylogenetic tree hinted towards a close relationship between the PGIPs of Pharsalusvulgaris (Kidney Bean) and that of Pisum sativum (Pea).
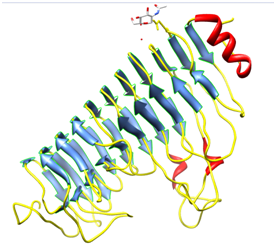
PGIP protein domain analysis: PGIPs belong to the Leucine-rich Repeat (LRR) super family of proteins containing tandem repeats of a 20-30 amino acid stretch of the extra Cytoplasmic type with a consensus that bear a conserved part [LxxLxLxxNxL or LxxLxLxxCxxL (L=I, L, V, F; N=N, T, S, C; C=C, S; x=any amino acid)] and a variable part. The LRR is believed to be specialized in protein-protein interaction. The LRR regions were identified for their location using Pfam,20 initially, in the PvPGIP2. A UniProtKB search of PGIP protein yielded reviewed hits, also found during MSA, namely PGIPs from Pyrus domestica (Pear), Oryza sativa (Rice), Arabidopsis thaliana (Mouse-ear cress) and Vitis vinifera (Grape). By using Ugene, annotation of LRR domain of PGIP proteins in the obtained plant species was studied. Further on, an MSA of LRR Domains of respective PGIPs was performed along with a guide tree analysis. A pair wise sequence comparison between PvPGIP2 and PsPGIP1 & PvPGIP1 and PsPGIP1 was also made.
PGIP structure analysis
Homology protein modelling: The structure of PvPGIP2 is downloadable from PDB where as structures for PvPGIP1 and PsPGIP1 were modelled using SWISS-MODEL (automated mode). Owing to high sequence similarity of PvPGIP2 with both PvPGIP1 and PsPGIP1, PvPGIP2 was used as template structure for modelling the two structures. The program automatically generates a model for the target along with structure evaluation results on the basis of Qmean Score and ANOLEA values. The further assessment for appropriateness of predicted structures was supplemented with exploration by Ramachandran plots.
Protein binding pocket analysis: Computed Atlas of Surface Topography of proteins (CASTp) provides an online resource for locating, delineating and measuring concave surface regions on three-dimensional structures of proteins. These include pockets located on protein surfaces and voids buried in the interior of proteins. The measurement includes the area and volume of pocket or void by solvent accessible surface model (Richards’ surface) and by molecular surface model (Connolly’s surface). CASTp can be used to study surface features and functional regions of proteins. The putative binding sites for the fungal enzyme FmPG were analyzed and predicted with the help of CASTp.20
Protein-protein docking: The protein docking software GRAMM-X, a web interface, uses GRAMM Fast Fourier Transformation methodology by employing smoothed Lenard-Jones potentials, refinement stage and knowledge based scoring to predict the docked complexes. An important feature of GRAMM is the ability to smooth the protein surface representation to account for possible conformational change upon binding within the rigid body docking approach. The web server frees users from complex installation of database-dependent parallel software and maintaining large hardware resources needed for protein docking simulations. A simple web interface of GRAMM-X accepts two PDB protein structures from the user, forms a job request and submits it to the execution queue on the cluster. Simulation results can be obtained on the server or through an Email notification from the web server according to the Lenard-Jones potential scoring function. By fixing FmPG (enzyme) as the receptor and the PGIPs (inhibitor) as ligand, the three pairs of proteins, namely FmPG-PvPGIP2, FmPG-PvPGIP1 & FmPG-PsPGIP1, were docked against each other. The output PDB file contains 10 models ranked as the most probable prediction candidates according to the Lenard-Jones potential scoring function.21
Protein- protein amino acid interaction
Analyses of atomic interactions in tertiary structures of proteins contribute richly to our understanding of sequence–structure relationships, structural basis of protein stability and protein evolution. Thus interactions within a protein structure and interactions between proteins in an assembly are essential considerations in understanding molecular basis of stability and functions of proteins and their complexes. Different kinds of interactions have been noted in the stabilization of tertiary structures, quaternary structures and assemblies of proteins. The KFC Server is a web-based implementation of the KFC (Knowledge-based FADE and Contacts) model-a machine learning approach for the prediction of binding hot spots, or the subset of residues that account for most of a protein interface’s binding free energy. The server facilitates the automated analysis of a user submitted protein-protein or protein-DNA interface and the visualization of its hot spot predictions. After the computational analysis, the user can visualize the results using an interactive job viewer able to quickly highlight predicted hot spots and surrounding structural features within the protein structure.22 The KFC model is comprised of two decision tree-based classifiers: K-FADE [based on shape specificity features calculated by the Fast Atomic Density Evaluator, or FADE] and K-CON (based on biochemical contact features). The three pairs of proteins, namely FmPG-PvPGIP2, FmPG-PvPGIP1 & FmPG-PsPGIP1 were hence analysed for putative hot spots. Protein Interactions Calculator (PIC) is a server which, given the coordinate set of 3D structure of a protein or an assembly, computes various interactions such as disulphide bonds, interactions between hydrophobic residues, ionic interactions, hydrogen bonds, aromatic- aromatic interactions, aromatic–sulphur interactions and cation-π interactions within a protein or between proteins in a complex. PIC server23 accepts atomic coordinate set of a protein structure in the standard Protein Data Bank (PDB) format. An option is provided, using RasMol interface and Jmol interface, for enabling visualization of structure in the graphics with interactions highlighted. It is possible to get the results by e-mail. It is also possible to download the output files. Hydrophobic and Ionic interactions are calculated for the three docked complexes, FmPG-PvPGIP2, FmPG-PvPGIP1 & FmPG-PsPGIP1.
Retrieval of information
The crystallized structure and FASTA sequence of Pharsalus vulgaris PvPGIP2 (PDB ID: 1OGQ) and Endo PolyGalacturonase enzyme Fusarium moniliforme FmPG (1HG8) is retrieved from PDB.
Sequence analysis
BLAST search carried out in order to locate sequence homolog’s from various plant species using ungapped alignment so as to include LRR motif during alignment. 28 sequences with minimum sequence identity of 50% and E-values >0.1 from various plant species were selected and analysed by MSA with CLUSTALW. A guide tree of the MSA generated on the basis of nearest neighbour approach depicts that Glycine max (Soyabean) and Pisum sativum (Pea) are close relatives of Pharsalu svulgaris (Kidney Bean). From literature studies it is already known that GmPGIP3 is effective in evading infection caused by Fusarium moniliforme by blocking its PG.24 Whereas in Pea the PGIP1 is able to prevent cyst nematode infection19 but its efficacy against FmPG is unknown.
Domain analysis
UniProtKB search revealed PGIPs from Pyrus domestica (Pear), Oryza sativa (Rice), Arabidopsis thaliana (Mouse-ear cress) and Vitis vinifera (Grape), also found during MSA, as reviewed entries with annotated LRR regions. Followed by a Pfam analysis of PvPGIP2 disclosing the location of LRR regions in the sequence. Location of LRR regions of PGIP from Pea was unknown.
Manual LRR sequence annotation
Annotation of the LRR regions in PGIPs from Pharsalusvulgaris (PGIP2 and PGIP1), Pyrus domestica (Pear), Oryza sativa (Rice), Arabidopsis thaliana (Mouse-ear cress) and Vitis vinifera (Grape) studied using UGENE. PGIP1 from Pisum sativum (Pea) annotated manually for its LRR region from knowledge based on repeats present in other PGIPs. Relationship between the PGIP proteins from Kidney bean and Pea established by MSA and NJ guide tree unvieled the plants as close relatives belonging to leguminous group. Relationship in terms of their LRR Domains evaluated by multiple alignment of the LRR regions from all the above annotated PGIP proteins points towards a crucial hint about the same characteristic function of PsPGIP1 (Pisum sativum) and PvPGIP as the LRR domains play the role in protein interactions and blocking the PGs from Fusarium moniliforme combating the infection. NJ guide tree confirms PsPGIP1 as a close relative of PGIP1 and PGIP2 from Kidney Bean. Pairwise sequence comaprizon of PsPGIP1 with PvPGIP1 and PvPGIP2 yields PsPGIP1 has 70% sequence identity with PvPGIP2 and 65% sequence identity with PvPGIP1. The question still to be answered is that will PsPGIP1 be able to inhibit FmPG or will be ineffective in blocking the enzyme?
Structure analysis
The crystal structure of PvPGIP2 being available at PDB (1OGQ) and due to high sequence similarity of PvPGIP1 and PsPGIP1 with PVPGIP2, the structures are modelled with SWISS-MODEL using PvPGIP2 as template.
Binding pocket analysis
Further, the analysis of putative binding sites for FmPG by CASTp brings into light the residues present in the binding pocket of the enzyme (Table 1).
Docking analysis
The flexibility of proteins in 3-D space poses a problem in protein-protein docking since both the receptor and ligand are flexible. The protein docking software GRAMM-X, a web interface, uses GRAMM Fast Fourier Transformation methodology by employing smoothed Lenard-Jones potentials, refinement stage and knowledge based scoring to predict the docked complexes. By fixing FmPG (enzyme) as the receptor and the PGIPs (inhibitor) as ligand, the three pairs of proteins are docked against each other. The output PDB file contains 10 models ranked as the most probable prediction candidates according to the Lenard-Jones potential scoring function
FmPG-PvPGIP2 complex
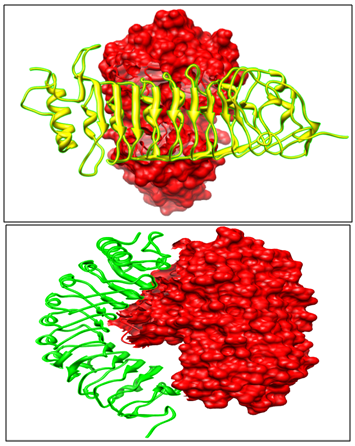
The experimental and literature studies,25 point towards the fact that PvPGIP2 blocks the activity of fungal enzyme FmPG. Out of the top ten models returned by GRAMM-X, selected model is such that the active site of FmPG is blocked by PvPGIP2 thus depicting enzyme inhibition (Figure 3).
FmPG-PvPGIP1 complex
Docking results indicate that PvPGIP1 is unable to block the active cleft of FmPG, thus failing to inhibit the enzyme (Figure 3).
FmPG-PsPGIP1 complex
When FmPG is docked with PsPGIP1 same orientation of the Pea PGIP protein with FmPG is observed like seen with PvPGIP2 from Kidney bean (Figure 4). Thus PsPGIP1 also blocks the active cleft of FmPG in the same way as done by the PvPGIP2. Hence we qualitatively observe that PsPGIP1 from Pea is capable of blocking the Fusarium moniliforme infection.

Protein-protein interaction analysis
KFC Server: It is based on a machine learning approach for the prediction of binding hot spots, or the subset of residues that account for most of a protein interface’s binding free energy. The KFC Server characterizes the local structural environment, compares that environment to the environments of experimentally determined hot spots and predicts if the interface residue is a hot spot (Figure 5). Quantitatively, the analysis of the interacting residues of FmPG with each of the three PGIPs namely PvPGIP1,PvPGIP2 from Kidney Bean and PsPGIP1 from Pea (Table 2) show that the same residues of FmPG are bocked by PvPGIP2 and PsPGIP1 and these residues belong to the active site as predicted by CASTp (Table 1). Therefore we infer that the fungal enzyme FmPG is being blocked by PsPGIP1 in the similar way as that by PvPGIP2.
Residues in Binding Pocket Predicted by Castp |
Residue Name and Number |
Cavity 1(green) |
HIS128, HIS 152, ASP 155, ILE156, THR157, 188HIS, 189ASN,191ASP,194ASP, ASP212,GLY234, ASN304,PRO307 |
Cavity 2(yellow) |
GLN96, LEU116, ASP121, SER124, THR133, ASP 140, ASP145, PRO148, ALA150 |
Cavity 3(purple) |
SER288, VAL290, SER292, THR293, THR294, PRO299, ILE300, ASN302, |
Cavity 4(orange) |
SER331,SER332, THR 340, LEU348, HIS352, THR354, ASP356, LYS360 |
Table 1 List of residues in the predicted binding pockets of FmPG
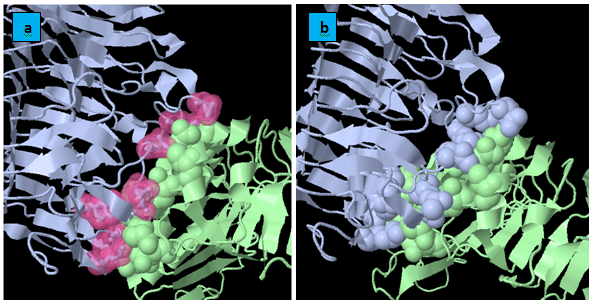
PIC: Protein interaction calculator: It is known that hydrophobic interactions and ionic interactions play a significant role in protein-protein interactions involving PG and PGIP. Thus here calculation of hydrophobic and ionic interaction are quantitatively analyzed for all the three docked complexes involving FmPG and PGIP proteins from Pharsalus vulgaris (PvPGIP1 and PvPGIP2) and Pisum sativum (PsPGIP1). Since from the docking results and the KFC server results it has been validated that PsPGIP1 is able to block the active site of the fungal enzyme FmPG. At this moment we are trying to cross validate the results on the basis of interactions involved in the complexes. On comparatively analyzing the interactions it was seen that similar residues of FmPG were involved in hydrophobic and ionic interactions in FmPG-PvPGIP2 and FmPG-PsPGIP1 docked complexes (Table 3).
Fm-PG- Pvpgip1 |
Fm-PG- Pvpgip2 |
Fm-PG- Pspgip1 |
|||
|---|---|---|---|---|---|
Residue Name |
Residue No. |
Residue Name |
Residue No. |
Residue Name |
Residue No. |
GLN |
51 |
LYS |
116 |
LEU |
303 |
PRO |
53 |
ASN |
121 |
ASN |
121 |
THR |
76 |
SER |
183 |
SER |
183 |
ASP |
106 |
LEU |
184 |
LEU |
184 |
ASN |
108 |
SER |
292 |
SER |
292 |
GLN |
110 |
THR |
293 |
THR |
293 |
GLN |
147 |
THR |
294 |
THR |
294 |
PRO |
177 |
ASN |
304 |
ASN |
304 |
ALA |
179 |
PRO |
307 |
PRO |
307 |
LYS |
180 |
THR |
308 |
THR |
308 |
SER |
183 |
SER |
331 |
SER |
331 |
LEU |
184 |
SER |
332 |
SER |
332 |
SER |
331 |
LYS |
360 |
LYS |
360 |
Table 2 Showing the residue name and residue number in the three docked complexes
Interactions |
FmPG |
PvPGIP2 (Kidney Bean) |
||
|---|---|---|---|---|
|
Residue No. |
Residue Name |
Residue No. |
Residue Name |
Hydrophobic interactions |
177 |
PRO |
95 |
PRO |
184 |
LEU |
116 |
ALA |
|
184 |
LEU |
118 |
PRO |
|
Ionic interactions |
188 |
HIS |
230 |
ASP |
Table 3 Showing the residue name and residue number protein interaction
The above study ascertains that a high level of sequence similarity not necessarily confers the proteins same functional characteristics, sometimes the structural fold and the hydrophobic as well as ionic interactions determine the functionality of the proteins. PsPGIP1 of Pisum sativum is a sequence homolog of PvPGIP2 & PvPGIP1 proteins of Pharsalus vulgaris and contains the conserved LRR region (LxxLxLxxNxL or LxxLxLxxCxxL) but its efficacy towards FmPG is unknown. Docking analyses suggest that the mode of binding of PsPGIP1 to the fungal enzyme FmPG is analogous to that of PvPGIP2 establishing that PsPGIP1 is able to block the active cleft of FmPG therefore being able to inhibit the fungal enzyme. Although PVPGIP1 and PvPGIP2 share a high sequence similarity yet their binding pattern to FmPG is entirely different from one another and thus PvPGIP1 fails to inhibit the enzyme. Protein-protein interactions study unveils the fact the residues of FmPG involved in interaction (hydrophobic & ionic) between the two docked complexes (i.e. PvPGIP1-FmPG complex & PsPGIP1-FmPG complex) are exactly same and belong to the active site of the enzyme. We hereby conclude from our studies that PsPGIP1 is effective in blocking the enzyme activity of PG from Fusarium moniliformin.
The authors acknowledge the Centre of Systems Biology and Bioinformatics, Panjab University, Chandigarh, India. Special thanks to Panjab University for providing facilities and guidance to carry out studies.
The author declares no conflict of interest.

©2017 Sood, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.