MOJ
eISSN: 2374-6912


Review Article Volume 1 Issue 1
State Key Laboratory of Reproductive Biology, Chinese Academy of Sciences, China
Correspondence: Yu Wang,State Key Laboratory of Reproductive Biology, Institute of Zoology, Chinese Academy of Sciences, Beijing 100101, China, Tel +861082619461
Received: May 29, 2014 | Published: June 10, 2014
Citation: Zhao Y, Ying Y, Wang Y. Developing crispr/cas9 technologies for research and medicine. MOJ Cell Sci Rep. 2014;1(1):20-25. DOI: 10.15406/mojcsr.2014.01.00006
CRISPR/Cas, comprised of Clustered regularly interspaced short palindromic repeats (CRISPR) and associated proteins (Cas), provides adaptive immunity against foreign DNA in bacteria and archaea. Type II CRISPR/Cas9 has attracted considerable interest as a tool to probe and manipulate biological systems. It has been engineered to introduce genome editing in a simple, flexible, and efficient manner. Various approaches have been explored to reduce the off-target effects, which limit its applications in certain circumstances. Moreover, a catalytically inactive Cas9 was used to guide various effectors and a labeling agent to specific DNA targets through protein fusion, thus enabling genetic/epigenetic regulation and visualizing specific genomic loci in living cells. Further, CRISPR/Cas9 could have a significant impact on medicine by facilitating the generation of cell lines and animal models for therapeutic screening and evaluation, and by providing a new promising avenue for gene therapy.
CRISPR/Cas9 technologies are being rapidly developed in multiple directions, enabling new approaches for biological research. In addition, they may transform how drugs are developed and how human patients are treated in the future. Here we review CRISPR/Cas9 technologies originated, their applications utilizing the natural function, expansion of applications by repurposing to other functions, efforts for precision improvement, and finally how CRISPR/Cas9 may impact medicine.
Keywords: crispr/cas9, genome editing, genetic modulation, epigenetic modulation, genomic dna labeling, drug screening, animal models, gene therapy
CRRNA, crispr rna; TRACRRNA, trans-activating crrna; PAMs, protospacer adjacent motifs; HDR, homology-directed repair; DSB, double-strand breaks; ZF, zinc fingers; TALE, transcription activator-like effectors; ZFN, zinc-finger nuclease; NHEJ, non-homologous end-joining; dCas9, dead” cas9; CRISPRi, crispr inference; sgRNA, single guide rna
The CRISPR/Cas system is immune defense machinery in bacteria and archaea that utilizes short RNAs to direct degradation of invading foreign nucleic acids, such as those from viruses or plasmids.1–3 This system essentially contains clustered regularly interspaced short palindromic repeats (CRISPRs) and their associated genes (Cas).2,4–7 CRISPR locus contains a series of short repeated sequences that are separated by non-repetitive spacer sequences derived from foreign genetic elements (Figure 1). This conserved repeat-spacer-repeat architecture was originally observed in the Escherichia coli genome in 1987.8 It was later found that the spacer sequences in CRISPR loci are identical to sequences in bacteriophage (phage) genomes and plasmids in 2005.9–11 More than 40 different Cas proteins have been reported so far.12 Based on the sequences and structures of Cas protein, CRISPR/Cas system is primarily classified into three types I, II and III.6 In CRISPR/Cas system, invading foreign DNA is processed by Cas nuclease into small DNA fragments, which are then incorporated into CRISPR locus of host genomes as the spacers. In response to repeat viral / phage infections, the spacers are used as transcriptional templates for producing CRISPR RNA (crRNA), which guides Cas to target and cleave DNA sequences of invading viruses and phages13 (Figure 1). A unique feature of Class II is that a single Cas protein performs multiple functions whereas in other classes multiple Cas proteins are required to orchestrate and fulfill the role.13–16 Such simplicity makes it an ideal system as a biomedical research tool.
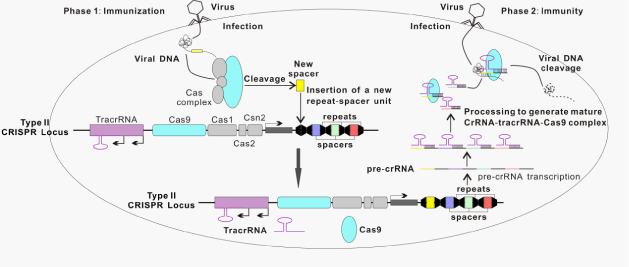
The type II CRISPR/Cas9 system is comprised of a long pre-crRNA transcribed from the spacer-repeat CRISPR locus, the multifunctional Cas9 protein, and a trans-activating crRNA (tracrRNA), which is important for processing the pre-crRNA and formation of the Cas9 complex. TracrRNAs first hybridize to repeat regions of the pre-crRNA. Endogenous RNase III cleaves the hybridized crRNA-tracrRNAs, and an unknown mechanism removes the 5′ end of sequence in each spacer, yielding mature crRNAs that remain associated with both the tracrRNA and Cas9. Finally, the mature complex surrogates the foreign DNA and locates a target sequence and cuts both strands.17 The target sequence matches the protospacer and is adjacent to short sequences known as protospacer adjacent motifs (PAMs) at the 3′ end.14,16 The PAM is an essential targeting component that serves as a self versus non-self recognition system to prevent the CRISPR locus itself from being targeted.18,19 The most commonly engineered system thus far, that of Streptococcus pyogenes, specifically recognizes the PAM sequence of NGG, where N can be any nucleotide.14,17,20
The ability of the CRISPR/Cas9 system to specifically target genomic sequence within living cells and organisms holds strong promise both as a powerful tool for biological research and as a potential avenue for gene therapy.21 For decades, traditional methods of gene targeting relied on homology-directed repair (HDR), a natural mechanism that cells use to repair double-strand breaks (DSB) in DNA. Mario Capecchi, Martin Evans and Oliver Smithies were awarded the Nobel Prize in 2007 for development of this technique.22 However, homologous recombination is often very inefficient without a DSB, thus targeting is always labor intensive with minimal success. To solve this problem, a DSB may be introduced at the targeting site, which triggers the recruitment of DNA repair machinery to the site, thus dramatically enhancing targeting efficiency.23,24
High specificity in the context of a genomic scale, ideally unique occurrence in the genome, needs to be achieved for precision in targeting. Most nucleases do not bind sequences that are long enough to reach high enough specificity, except a class named meganucleases. Therefore, meganucleases were engineered to facilitate genome editing via cutting DNA in a highly specific and targeted manner.25–27 Fusion constructs of non-specific nuclease and repetitive domains that confer DNA recognition specificity, including zinc fingers (ZF) and transcription activator-like effectors (TALE), have been developed and used in genome engineering.21 For example, the CCR5 gene was modified by a zinc-finger nuclease (ZFN) in CD4 T cells of human AIDS patients, which are then infused autologously to decrease the level of HIV infection.28 While all these are protein-based systems, which require significant effort in assembling the constructs, CRISPR/Cas9 system is simpler, more flexible, effective and confers DNA recognition specificity by RNA (Table 1).
CRISPR/Cas9 |
TALE |
ZF |
Mega nuclease |
|
|---|---|---|---|---|
Type of recognition |
RNA-DNA |
Protein-DNA |
Protein-DNA |
Protein-DNA |
Off-target effects |
More potential off-target effects than TALEs, ZFs and Mega nuclease |
Less observed off-target effects than CRISPR/Cas9 |
More potential off-target effects than TALEs |
Potential off-target effects |
Multiplexing |
Capable and easy |
Labor intensive and Rarely used |
Labor intensive and Rarely used |
Labor intensive and Rarely used |
Target sequence |
Adjacent to PAM, a short sequence varies among CRISPR/Cas9s from different species |
Each TAL repeat binds a base pair of DNA. Sequences targeted by TAL effector repeats are typically directly preceded by a thymine (T) that is required for maximal activity |
Each zinc finger binds a 3bp DNA target. Not all 3bp sequences can be targeted. Assembly of 3-4 zinc finger modules is required for specificity in recognition |
Only pre existing mega nuclease recognition site. The absence of that same site from the targeting vector is required |
Applications in research |
1. Genome editing ; |
1. Genome editing; |
1. Genome editing; |
1. Genome editing; |
Applications in clinic |
Gene editing of CCR5 in autologous CD4+T cells of persons infected with HIV |
Table 1 Comparison of CRISPR/Cas9, TALE, ZF and Mega nuclease technologies
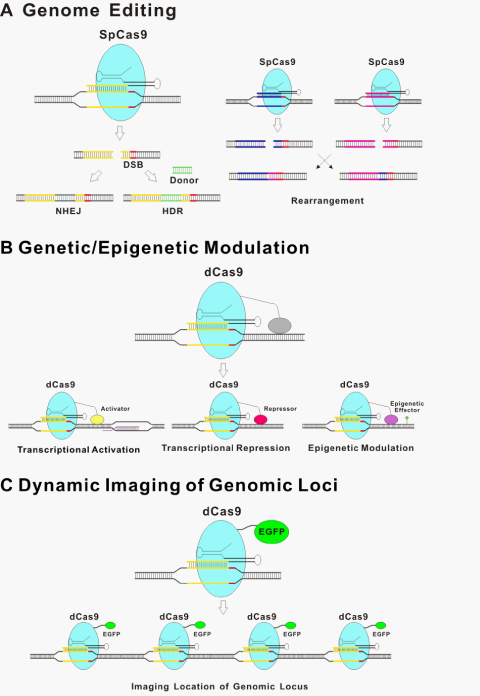
The underlining principle for all the new genome editing technologies is that the introduced DSBs are repaired by either non-homologous end-joining (NHEJ)29 without homologous DNA or more precise HDR in its presence30 (Figure 2A). NHEJ is an error-prone process that is often accompanied by insertion or deletion of nucleotides (indels) at the targeted site, sometimes resulting in a genetic knockout of the targeted region of the genome due to frameshift mutations or the insertion of a premature stop codon.31 Moreover, Cas9-induced DNA breaks can promote efficient rearrangement between pairs of targeted loci over a long distance through the non-homologous end-joining (NHEJ) pathway of DSB repair.32 Cas9 has been used to modify genome sequence through NHEJ in bacteria,33 in cultured human cancer cell lines and human induced pluripotent stem cells,34–37 as well as in whole organisms, including zebrafish, mouse, and monkey.38–43 In the presence of homology DNA of the target site, HDR also occurs so that host genome DNA between homologous arms is exchanged out and donor DNA is inserted. Such approach has been used for genome engineering with high precision, from introducing single nucleotide mutations to introducing new genes to specific locations in the genome.31,44 As an example, two basepair substitutions of Tet genes can be introduced into the mouse genome via zygote microinjection of Cas9 mRNA, gRNA, and oligo donors.38
Following the initial demonstrations that type II CRISPR/Cas9 could be programmed to cut various DNA sites in bacteria,14 a series of reports subsequently showed that CRISPR/Cas9 systems can be engineered to introduce genome editing in a wide variety of species including crop, zebrafish, mouse, rat, rabbit, pig and monkey.38–43,45–50 Moreover, CRISPR/Cas9 based genome editing can be easily multiplexed by introducing multiple gRNAs.35,38,50,51 Perhaps the best example for utilizing this great flexibility is the functional genomics screens with a large pool of gRNAs (Figure 2A).52–55
Although CRISPR/Cas9 demonstrated obvious advantages for genomic modifications in biology, high-frequency off-target effects are the bottleneck for some applications.56–59 Off-target cleavages are one major concern in CRISPR/Cas9-mediated genome editing. The repair of off-target DSBs could result in deletions, inversions, translocations, and unknown off-target mutations, which may lead to undesired activation of oncogenes or inactivation of tumor suppressor genes. Several studies investigated the patterns of off-targeting effects and discovered that mismatches are generally better tolerated at the 5′ end of the 20-nt targeting region of the gRNA than at the 3′ end.14,32,34 The 8-12bp at the 3′ end of the targeting sequence, known as the ‘seed’ sequence, are crucial for target recognition in vitro and in bacterial cells.14,33,35,60,61 However, the effects of mismatches are not always predictable based on their location within the gRNA targeting region; some mismatches in the 5′ end may have dramatic effects, whereas some in the 3′ end do not obviously affect CRISPR/Cas9 activity.56 In addition, not all nucleotide substitutions at a given position necessarily confer equivalent effects on activity.62
Various approaches were explored to reduce the off-target mutagenic effects of CRISPR/Cas9. One strategy is to use paired “nickases”. Cas9 variants that cut one strand rather than both strands of the target DNA sites known as “nickases”. Because individual nicks in the genome are repaired with high fidelity, simultaneous nicking via appropriately offset guide RNAs is required for double-stranded breaks and extends the number of specifically recognized bases for target cleavage. The paired nickases targeted to sites on opposite DNA strands separated by 4 to 100bp can efficiently introduce both indel mutations and HDR events with a single-stranded DNA oligonucleotide donor template in mammalian cells.57,59,63,64 Another proposed approach for reducing Cas9-induced off-target effects of gRNAs in human cells involves the use of truncated gRNAs.65 These truncated gRNAs with a shortened 5′ end are 17 or 18 nucleotide long. They generally function as efficiently as full-length gRNAs in directing on-target Cas9 activity but show decreased undesired mutagenic effects at off-target sites by 5,000-fold or more.65 Recently, an approach was also used to improve DNA cleavage specificity by fusions of catalytically inactive Cas9 and FokI nuclease (fCas9).66 Two fCas9 monomers simultaneously bind target sites 15 or 25 base pairs apart. FokI nuclease cleaves DNA only if the target sites are occupied simultaneously by two FokI domains at a specified distance and in a specific half-site orientation. It was reported that fCas9 modified target DNA sites with >140-fold higher specificity than wild-type Cas9 and with an efficiency similar to that of paired Cas9 'nickases'.66
Genetic/epigenetic modulation
The CRISPR/Cas9 system has the potential to regulate endogenous gene expression. Catalytically inactive or “dead” Cas9 (dCas9) bearing mutations at the two cleavage sites renders Cas9 unable to cut DNA, but it can still be recruited by gRNAs to target specific DNA sites within the genome.14,21,67 Therefore, dCas9 with heterologous effector domain functions can be recruited to specific genome loci, thus modulating the genetic/epigenetic states (Figure 2B). dCas9 fusions to a transcriptional activation domain (e.g. VP64 and the p65 subunit of nuclear factor kappa B; NF-κB) or a transcriptional repression domain (e.g. the Krüppel-associated box (KRAB) domain) implements precise and stable transcriptional control of target genes.68–72 For example, single or multiple gRNAs can direct a VP64 transcriptional activation domain fused with a dCas9 protein to activate the expression of several endogenous genes.68,69 An alternative strategy of using aptamer fused with gRNA for binding an effector domain has also been reported.64 Although activity with this strategy is less robust than direct fusions to dCas9, this type of configuration might provide additional options and flexibility for recruitment of multiple effector domains.21 Interestingly, a synergistic effect of multiple gRNAs to target the same promoter for more effective modulation was observed in multiple studies.64,68,69,71
A modified CRISPR/Cas9 system named CRISPR inference (CRISPRi) has been developed for regulation of gene expression in eukaryotic cells.73 The CRISPRi system required that a catalytically inactive Cas9 protein was co-expressed with a customizable single guide RNA (sgRNA) to form a recognition complex, which interferes with transcriptional elongation and the binding of RNA polymerase and transcription factor. However, the degree of repression achieved by CRISPRi is modest in mammalian cells.73
Recently, several laboratories reported that epigenetic modifications of DNA and chromatin can be introduced via fusing epigenetic effectors with TAL repeats.70,74,75 A dCas9 fusion might also be used to perform targeted ‘epigenetic modulation’. Such targeted perturbation of the epigenetic status of specific loci would allow, for the first time, functional dissection of causative events among candidates found in association studies.
Dynamic imaging of genomic loci
d Cas9 fused with a fluorescent protein have been used to visualize specific genome loci in live cells (Figure 2C).76 The movements of telomeres labeled by dCas9::GFP or TRF1, a major telomere-binding protein, were characterized. No significant difference was observed between these two labeling methods, thus supporting dCas9 as a neutral probe of genomic DNA. This labeling strategy, among others, provides a useful tool for studying chromosome dynamics and structure and further extends dCas9-based applications.
Potential clinical applications of crispr/cas9
Given the tremendous value of CRISPR/Cas9 as a versatile tool to modulate various aspects of biological systems, it may provide equally exciting opportunities in clinical applications in addition to those in basic biological research.
There are two avenues ahead that CRISPR/Cas9 technologies could impact clinical translation. First, CRISPR/Cas9 could dramatically accelerate the pace for generating cell lines and animal models that harbor desired disease genes and reporters,41,42,48–50,77,78 which are used for drug screening and preclinical evaluation. CRISPR/Cas9 would be even more empowering when partnered with other transforming technologies such as iPS cell reprogramming and haploid stem cell targeting.49,79–87 Second, CRISPR/Cas9 itself might provide a new avenue for gene therapy. The success of ZFN in treating AIDS in a human clinical trial brought a lot of excitement and hope for treating a wide-variety of human diseases using similar approaches including CRISPR/Cas9.28 Considering the versatility of CRISPR/Cas9 systems, one can envision multiple ways for it to be applied in clinic one day, including genetic correction,42,77 epigenetic correction, and modulation of “disease” gene expression.78
To make the utility of CRISPR/Cas9 a reality in clinic, scientists first need to engineer the CRISPR/Cas9 system to a much higher level. First, precision of this technology needs to be further optimized and off-target effects need to be eliminated or controlled in order to ensure safety. Second, effective delivery of the CRISPR/Cas9 components, especially using non-integrative delivery technologies, shall be developed and matured. Third, the versatility of Cas9 to target any DNA sequence would need to be achieved which rely on the discovery of more orthogonal systems from nature or the development of Cas9 variants in laboratories that recognize distinct PAMs.88
Work in Y.W.’s laboratory is supported by the National Basic Research Program of China (Grant No. 2014CB964900) and funding from the Chinese Academy of Sciences.
The author declares no conflict of interest.

©2014 Zhao, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.