MOJ
eISSN: 2471-139X


Mini Review Volume 2 Issue 3
1Department of Cell Biology and Anatomy, Rosalind Franklin University of Medicine and Science, USA
2Division of Biomedical Statistics and Informatics, USA
3Department of Biomedical Sciences, Rosalind Franklin University of Medicine and Science, USA
Correspondence: Michael Sarras P Jr, Department of Cell Biology and Anatomy, Chicago Medical School, Rosalind Franklin University of Medicine and Science, 3333 Green Bay Road, North Chicago, IL, 60064, USA, Fax 18475783432
Received: April 05, 2016 | Published: April 28, 2016
Citation: Sarras Jr MP, Leontovich AA, Intine VR. A potential GDNA Methylation epigenetic mechanism to explain the long term complications of type 1 and 2 diabetes. MOJ Anat Physiol. 2016;2(3):89–92. DOI: 10.15406/mojap.2016.02.00049
Metabolic memory (MM) is the persistence of diabetic (DM) complications even after glycemic-control is pharmacologically achieved. Using a zebrafish diabetic model that induces a MM state, we previously reported that in this model, tissue dysfunction was of a heritable nature based on cell proliferation studies in limb tissue and this correlated with epigenetic gDNA-methylation changes that paralleled alterations in gene expression. Bioinformatics analysis found that gene expression of the DNA replication/DNA metabolism process group was altered in the DM state and altered expression continued into MM state. In this mini-review, we propose that the underlying mechanism(s) that explain how these gene expression changes relate to the long term complications observed in diabetes stems from alterations in the ability of transcription factors to bind to their respective gDNA binding sites. gDNA methylation changes of promoter regions (both proximal and distal to the transcription start site) would disrupt the ability of transcription factors to bind to their gDNA binding sites and this would be tissue specific; thus explaining the variety of problems observed patients suffering from the long term complications of diabetes mellitus (e.g. problems associated with the kidney, retina, skin wound healing processes, and angiogenesis, etc.). Clinical studies are now in planning for the translation of these findings to human DM.
Keywords: diabetes, metabolic memory, zebrafish, epigenetics, gdna methylation, bioinformatics, transcription factor binding
MM, metabolic memory; DM, diabetes mellitus; STZ, streptozocin; TSS, transcription start site; MRs, methylated regions
Current research points to hyperglycemia-induced changes to the methylation of gDNA as a contributing factor in the transition from a normal physiological state to one of diabetes mellitus.1‒14 In addition, persistence of this altered gDNA methylated state has been proposed to underlie the induction of changes in tissue-specific gene expression patterns that are associated with the many secondary complications observed in patients with both type-1 and type-2 diabetes.1‒14 The data, to date, are based on correlative findings but functional studies are required to establish a mechanistic link between: hyperglycemia, gDNA methylation changes, altered tissue-specific gene expression patterns, gene regulation systems affected by gDNA methylation, and the subsequent appearance of the secondary complications associated with diabetes mellitus. This mini-review will propose a hypothesis and specific experiments to test this hypothesis that will establish a functional link between the above mentioned pathogenic processes. The review begins with a review of the literature that points to the role of gDNA methylation in the induction of diabetes and its long term secondary complications, and will then. Propose mechanisms to explain, in part, these processes and present specific functional studies to test these proposed mechanisms.
Current data pointing to gDNA methylation as an underlying epigenetic mechanism(s) of the long term complications observed in diabetes
In addition to issues related to chronic glycemic dysregulation, a major problem with diabetes mellitus (DM) (both type 1 and 2) is its long term complications. In this regard, patients with diabetes encounter a multitude of tissue dysfunctions related to: cardiovascular disease, aberrant angiogenesis, retinopathy, nephropathy, neuropathy, and impaired wound healing.1‒14 Our laboratory has previously developed and reported on an adult zebrafish model of type 1 DM that has unique strengths for elucidating the mechanisms underlying the long term complications of the disease.2,13,14 In this model, streptozocin (STZ) induced hyperglycemia (serum glucose=315 +/- 40.96mg/dL) is accompanied by the full range of diabetic complications seen in patients with DM.13,14 Additionally, we have shown that withdrawal of STZ results in regeneration of pancreatic b-cells and the return of previously diabetic fish to a physiologically normal glycemic state within 2weeks (serum glucose=62.5 +/- 13.6mg/dL). However, in contrast, the tissue deficits associated with hyperglycemia persisted permanently (e.g. impairment of angiogenesis,15 impairment of skin wound healing13 and impairment of limb regeneration (fin regeneration) that results from an inhibition of cell division.13 Multiple controls indicated that this was a direct effect of b-cell necrosis-induced hyperglycemia and not due to any secondary effects of STZ treatment.13,14 These findings are consistent with large scale DM clinical trials whose results indicate that once initiated, diabetic complications persist and continue to progress unimpeded even when glycemic control is achieved through pharmaceutical intervention.1,5,7,16 Additionally, this persistence in diabetic tissue deficits has also been supported by multiple lines of experimental laboratory evidence3,6,10‒13 and collectively, this data indicate that the initial hyperglycemic period results in permanent abnormalities in the target organs. This harmful phenomenon has been termed, Metabolic Memory (MM)8,9 and to date, the underlying molecular mechanism(s) of MM remain unknown. Studies from our laboratory have provided evidence that epigenetic processes contribute to the long term deficits observed in the DM/MM zebrafish model and are likely a molecular component underlying metabolic memory in this model. Specifically, we have reported that hyperglycemia induces changes in gDNA methylation of specific genomic loci and these changes persist into the MM state following pancreatic b-cell regeneration when normal glycemic control is re-established systemically. As studied in the fin, control, DM and MM tissues were analyzed by MeDIP-sequencing and microarray techniques. Bioinformatics analysis of the data found that genes of the DNA replication/DNA metabolism process group (with up-regulation of the apex1, mcm2, mcm4, orc3, lig1, and dnmt1 genes) were altered in the DM state and these molecular changes continued into MM as impaired cell division in the fin were observed.2 Interestingly, persistent gDNA-methylation changes could be found as far as 6-13kb upstream of the transcription start site for these genes suggesting potential higher levels of epigenetic control.2 These gDNA methylation changes were accompanied by permanent alterations in mRNA expression patterns that correlated with the observed dysfunctions in the fin and in other tissues of the organism.
Potential gene regulation mechanisms underlying the long term complications observed in diabetes
Preliminary bioinformatics analyses indicate that both proximal and distal promoter regions (relative to each gene’s transcription start site [TSS]) have alterations in their gDNA methylation of cytosine bases (termed Methylated Regions, MRs). In this regard, we propose the following working hypothesis: “Persistent tissue dysfunction in the DM/MM states correlates with hyperglycemia-induced DNA methylation changes within transcription factor binding sites”. To test this hypothesis we will employ bioinformatics analysis of methylated genomic regions upstream and downstream of the TSS of those genes that had been previously identified to determine if they contained transcription factor binding sites that could be affected by gDNA methylation changes. As explained in our recent report,2 these studies will focus on genes of the zebrafish caudal fin because previous research has established that fin tissue is best suited for experimental creation of a “pure” metabolic memory tissue.2,13,14 Other tissues of the zebrafish (e.g. kidney, retina, skin, etc.) enter the MM state following b-cell regeneration, but unlike the fin; it is more difficult to form a tissue that lacks the residual molecules that were created in the original hyperglycemic state such as ROSs (Reactive Oxygen Species) and AGEs (Advanced Glycation Endproducts). The presence of such pathology-inducing molecules as ROSs and AGEs introduces complicating variables that makes evaluation of the epigenetic effects more difficult to interpret. Figure 1 depicts the generation of “pure” metabolic memory tissue in the zebrafish DM/MM model.
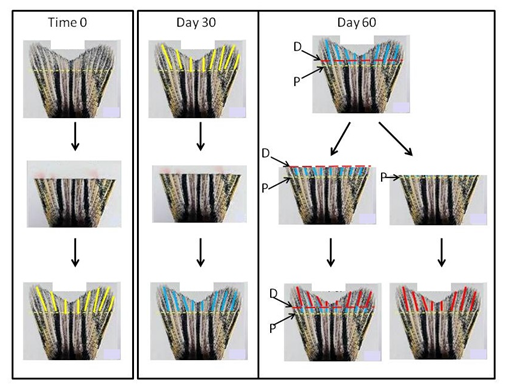
To validate these bioinformatics findings, we will establish that
Such an analysis will form a foundation for future studies to determine why particular gene regulatory regions are targeted for gDNA methylation changes following episodes of hyperglycemia. Clinical studies are now planned to translate these animal model findings to the human DM condition.
This review has described data pointing to gDNA methylation as an epigenetic mechanism that at least partially explains the long term complications observed in patients with type 1 or 2 diabetes. Future studies will elucidate these mechanisms and potentially lead to therapeutic approaches to prevent or reverse the long term complications of this disease. Table 1 summarizes the findings and proposals of this mini-review.
The studies described in this manuscript were supported to funds from the Iacocca Family Foundation, National Institutes of Health Grant (NIDDK) DK092721 (to RVI), and Rosalind Franklin University research funds awarded to MPS Jr.
Author declares that there is no conflict of interest.

©2016 Sarras, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.