Journal of
eISSN: 2475-5540


Research Article Volume 2 Issue 1
1Centre for Healthy Brain Ageing, University of New South Wales, Australia
2Department of Neurology, Hianjin Huanhu Hospital, China
Correspondence: hang P, Department of Neurology, Tianjin Huanhu Hospital, Tianjin, China, Tel 8.61E+12
Received: October 15, 2016 | Published: January 17, 2017
Citation: Kim J, Sachdev PS, Zhang P, et al. Stem cells-based therapeutics for parkinson’s disease - a transcriptomic analyses during dopaminergic neuron differentiation under 3- and 2- dimensional environments using human embryonic stem cells. J Stem Cell Res Ther. 2017;2(1):12-22. DOI: 10.15406/jsrt.2017.02.00052
Parkinson’s disease (PD) is a neurodegenerative disease which is caused by many factors including progressive degeneration of dopamine (DA)-secreting neurons which reside in the midbrain substantia nigra compacta (SNc). Current available treatments comprise of intake of DA replenishing drugs or implantation of electrical impulse device. However, the short-term effect of the treatments and risks of side effects have severely limited the widespread application of these therapies for all patients with PD. Hence, human embryonic stem cells (hESCs), which are capable of both self renewal and differentiation into all cell types of human body, could potentially provide a renewable source of surrogate DA neurons for transplantation into PD patients. One of the challenges in using hESCs therapeutically is the establishment of protocols that could effectively direct their differentiation into functional DA neurons. A specific investigation on the derivation of DA neurons was carried out by using a three-dimensional (3D) environment such as encapsulation. Characterization study by microarray was performed to analyze the global expression profile in 3D-derived DA neurons after 28 days of differentiation. In comparison to the samples of DA neuronal differentiated hESCs under 2D platform for 28 days, the analysis detected the reduced expression of gene that are involved in pluripotency or mitosis but increased expression of genes that are involved in neuronal developments such as Wnt, hedgehog and mitogen-activated protein kinase (MAPK) signaling pathway. The results suggest that the 3D differentiation system may have affected the regulatory or signalling mechanisms which enhanced the rate of differentiation towards ectoderm.
Keywords: neurodegenerative disease, neural stem cells, mesenchymal stem cells, cell replacement therapy
PD, parkinson’s disease; DA, dopamine; SNc, substantia nigra compacta; NSCs, neural stem cells; MSCs, mesenchymal stem cells; EST, expressed sequence tag; hESCs, human embryonic stem cells; SAGE, serial analysis of gene expression; MPSS, massively parallel signature sequencing; GO, gene ontology; PCA, principle component analysis; RMA, robust multichip average; BP, biological process; CC, cellular component; MF, molecular function
As described previously for potential cell replacement therapy for PD, many different types of cells have been considered, including neural stem cells (NSCs), ESCs, mesenchymal stem cells (MSCs) and trans differentiated cells.1,2 In each case, questions concerning character of the transplanted population, signals directing differentiation and specificity of differentiation have been raised. Therefore, there has been a great interest in understanding the molecular events/pathways involved in DA neuronal differentiation. Until recently, investigating developmental events that occur in DA neuronal differentiation has been difficult due to the lack of consensus over surrogate markers of efficacy and the availability and cost associated with assessing a reasonable number of markers. Multiple analytical techniques to assess gene expression in defined cell types have been developed, including microarray analysis, expressed sequence tag (EST) enumeration, serial analysis of gene expression (SAGE) and massively parallel signature sequencing (MPSS). Microarray analysis offers an advantage by allowing one to investigate thousands of genetic transcripts simultaneously with digital accuracy to particular cell populations or signalling pathways to assess the state of ESC differentiation.3,4 In addition, the application of the array allows rapid assessment of the state of a cell population that cannot be identified by simpler means e.g. morphology. Also, it can provide new insights into identification of specific markers of differentiation, drug development and developmental pathways. In this paper, a transcriptomic microarray was used to investigate the underlying molecular differences between hESCs differentiated under 2D and 3D systems.
The hypothesis for this study includes that the hESCs differentiated under 3D system will have higher expression levels of genes or pathways related to DA neuronal differentiation than the cells derived under the conventional 2D culture system. The main aims were to assess and identify the transcriptomic differences during differentiation of hESCs under 3D and 2D platform by carrying out:
Cell line classification
An embryonic stem (ES) cell line, E25 at various passages was cultured under different conditions were used to investigate differential gene expressions (Table 1). The E2 cell lines which were differentiated under 2D or 3D environment for 28 days as described previously that utilized inducers such as sonic hedgehog and fibroblasts growth factor 8a6 and were subjected to genome-wide transcriptomic analysis using Affymetrix Human Genechip® Gene 1.0 ST arrays. As a control comparison, E2 that was maintained in a pluripotent state was subjected to transcriptomic analysis. The passage numbers of each condition were chosen that showed normal karyotype (data not shown) and also we demonstrated previously that ES cells of different passages grown under feeder-free environment as in the present experiments exhibit minimal variation in global as well specific pluripotent gene expressions.7
Cell Lines |
Passage # |
Culture Condition |
Abbreviation |
# of Biological Replicates (n) |
E2 |
P23-p30 |
Undifferentiated |
UNDIFF/E2 |
3 |
E2 |
P35-37 |
Differentiated under 2D platform for 28 days |
DIFF/2D |
3 |
E2 |
P35-38 |
Differentiated under 3D platform for 28 days |
DIFF/3D |
2 |
Table 1 Classification of cell lines
The terminologies that are used throughout this paper are listed here along with each individual cell lines identity and the type of culture used.
Minimum information about a microarray experiment (MIAME)
Normalized data were submitted to Gene expression omnibus (GEO) archive according to MIAME recommendation (http://www.ncbi.nlm.nih.gov/geo/). A GEO accession number was assigned: GSE0000.
Microarray gene expression & data analysis
Affymetrix data array CEL result files were processed with Partek Genomic Suite v6.5 software (Partek). Independent comparisons via one-way analysis of variance (ANOVA) method were performed to identify differentially expressed genes for the following groups; (i) UNDIFF vs. DIFF (both 2D and 3D) and (ii) 2D vs 3D. False discovery rate (FDR, Benjamini-Hochberg) statistical method was used to counter multiple hypothesis testing which filters out the possibility of incorporating false positives. Differentially expressed genes were computed based on a FDR-corrected p-value < 0.05 and acquiring at least a 2-fold change to ensure downstream analysis was not affected by the presence of false positives.
Gene ontology enrichment
Gene ontology (GO) enrichment analysis was performed with Partek to identify over-represented GO-classes; molecular function, biological process and cellular components. Analysis was restricted to functional groups with more than 2 genes and statistics calculated using Fisher’s exact test. The enrichment score is a modified Fisher’s exact p-value and determines whether genes falling into a specific category differs by function and is used for gene enrichment.
Establishment of specific gene lists
Specific gene lists that are involved in the DA neural differentiation of hESCs were analysed which includes: neuronal ion channels, DA synthesis, Wnt, Sonic hedgehog (SHH), MAPK signaling and PD pathways. The gene lists (~55 genes from each pathway, minus the housekeeping genes) were obtained from Super Arrays Biosciences website: http://www.sabiosciences.com.
Generation of dendograms and heat maps
Heat maps were generated using the software, Multi Experiment viewer (MeV) v4.6.2 using the hierarchical clustering function (Saeedet al. 2006) of the mean signal intensity values for transcripts in each cell line. Hierarchical clusterings of specific gene lists were performed by complete linkage using Euclidean distance as a similarity metric. The scale bars represent an arbitrary genetic distance between cell lines.
Other analysis
Full data set of 33,297 transcript values were analysed; principle component analysis (PCA) was generated using Parktek, whereas scatter plots and global dendogram were generated using the software, Gene Spring GX v11.2 (Agilent Technologies). Scatter plots were represented by the absolute values of log2 transformed fold changes. This was determined by subtracting the mean signal intensity values of the cell type of interest against a control cell type e.g. pluripotent hESCs minus 3D differentiated hESCs. Correlation coefficients were calculated based on the mean and individual signal intensity values of relevant cell types using Microsoft Excel’s “CORREL” function.
Pathway analysis
Partek pathway function (Partek) was used to investigate if there is a significant enrichment of genes of interest in a particular pathway. Pathway enrichment analysis function was selected using Fisher’s exact test and restricted the analysis to pathways with more than 2 genes. The p-value is calculated using the hyper geometric distribution.
Global gene expression data of undifferentiated vs differentiated hESCs
A global transcriptomic analysis of the samples was performed; E2, 2D vs 3D. Using PCA, all of the transcript expression values were collated into a single data point and plotted accordingly to its most variable factors e.g. principle components between cell types. PCA revealed that UNDIFF clustered furthest away from the DIFF as expected (Figure 1). Interestingly, 3D was grouped away from 2D on the PC#3 axis, resulting 3 distinct components of separation; E2, 2D and 3D cell populations. The differential clustering profiles between 2D and 3D suggest that this may be caused by two different dimensional differentiation systems.
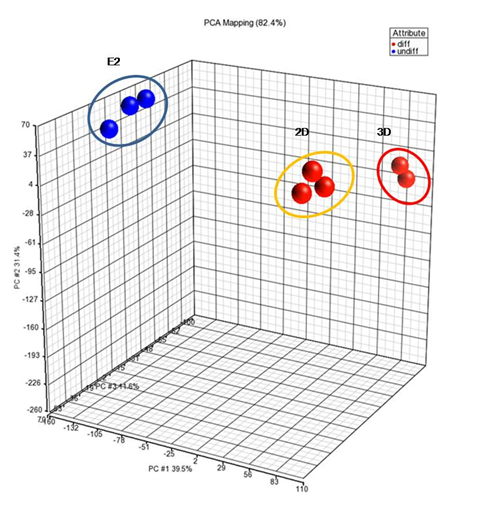
Figure 1 The correlation between UNDIFF and DIFF cells.
A 3D PCA-covariance plot with an overall principle component (PC) of 82.4%. Three distinctive groups were observed, E2 being clustered furthest away from DIFF groups (2D and 3D). Clustering is represented by colored spheres with the groups’ names.
Scatter plots are commonly used as another visual analysis tool for determining the overall correlation between two individual cell populations. Robust multichip average (RMA) normalized mean signal intensity values of each transcript were graphed for a visual representation of the similarity between two samples (Figure 2). Quantitatively, E2 and 3D had a lower correlation coefficient in comparison to E2 and 2D (r = 0.909501, r = 0.927886, respectively). Additionally, 3D and 2D were highly correlated (r = 0.95345) (Figure 2). The hierarchical clustering diagram illustrated that UNDIFF could be discriminated from the DIFF cell populations analyzed in this study, as expected (Figure 2). From all these data, it is suggested that the overall gene expression profile seen in UNDIFF could be discerned from that seen in DIFF.
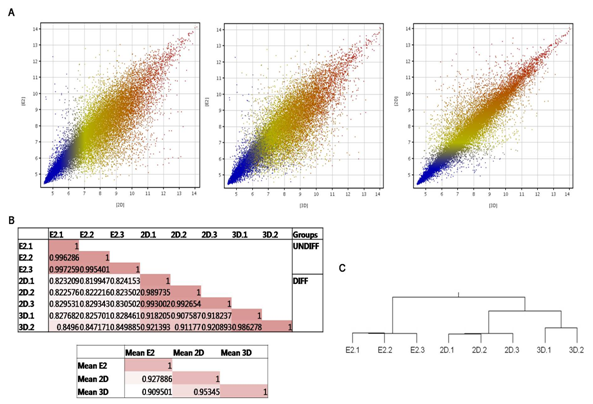
Figure 2 Global gene expression profiling. (A) Scatterplot representation of the mean signal intensity values of the 33,297 transcripts across different comparisons. (B) Correlation coefficients (r) were calculated using the mean signal intensity values for each cell line (including comparison of each of the biological replicates). An r-value 1 indicates perfect correlation and is represented by the darkest shading. Less correlated pairs are represented by lighter shades. (C) Global dendogram based on the overall gene expression profiles of the samples. Two distinct clusters corresponding to UNDIFF and DIFF, followed by further separation between 2D and 3D.
Differentially expressed genes in undifferentiated and differentiated hESCs
Followed by the global transcriptomic analysis of the samples, the differential gene expression profiles of the UNDIFF and DIFF were examined. Multiple hypothesis testing using FDR correction was used to narrow down candidate genes and identified the transcripts with a greater than 2-fold difference in normalized expression between the two samples. The analysis identified 9005 up-regulated transcripts in UNDIFF and 5461 up-regulated genes in DIFF.
A total of 14,466 genes were differentially expressed which included the widely accepted stem cell markers such as OCT3/4, teratocarcinoma-derived growth factor (TDGF1), NANOG and DNA methyl-transferase 3B (DNMT3B) were significantly down-regulated to 60.90, 319.14, 23.52 and 21.86 folds respectively, in DIFF against UNDIFF. The GO enrichment analysis of up-regulated genes in UNDIFF was found to be enriched in genes known or predicted to be involved in GO: 6397: mRNA processing, GO: 6396: RNA processing, GO: 6260: DNA replication, GO: 3899: DNA-directed RNA polymerase activity, GO: 34062: RNA polymerase activity (Table 2). This suggests that the high expression level of ribosomal genes in UNDIFF is in a state of active metabolism and growth or proliferation which are characteristic of stem cells.
Function UNDIFF up |
Type |
Enrichment Score |
Enrichment p-Value |
GO ID |
DNA replication |
BP |
7.53814 |
0.000532 |
6260 |
Cell proliferation |
BP |
5.64846 |
0.003523 |
8283 |
Nucleoplasm |
CC |
5.48496 |
0.004149 |
5654 |
DNA-directed RNA polymerase III complex |
CC |
3.76707 |
0.02312 |
5666 |
Regulation of DNA replication initiation |
BP |
3.43515 |
0.032221 |
30174 |
Regulation of transcription from RNA polymerase III promoter |
BP |
3.43515 |
0.032221 |
6359 |
Regulation of DNA replication |
BP |
3.0853 |
0.045716 |
6275 |
Function DIFF up |
Type |
Enrichment Score |
Enrichment p-Value |
GO ID |
Neuron recognition |
BP |
9.02714 |
0.00012 |
8038 |
Synaptic transmission |
BP |
7.2256 |
0.000728 |
7268 |
Neuron adhesion |
BP |
7.57188 |
0.000515 |
7158 |
Extracellular structure organization |
BP |
7.17824 |
0.000763 |
43062 |
Axon guidance |
BP |
4.75366 |
0.00862 |
7411 |
Synapse organization |
BP |
4.42452 |
0.01198 |
50808 |
Cell morphogenesis involved in differentiation |
BP |
4.15333 |
0.015712 |
904 |
Axon |
CC |
3.98271 |
0.018635 |
30424 |
Neuron projection |
CC |
3.50766 |
0.029967 |
43005 |
Brain development |
BP |
3.37537 |
0.034205 |
7420 |
Regulation of cell differentiation |
BP |
3.02045 |
0.048779 |
45595 |
Table 2 Differential gene expression of UNDIFF vs DIFF
GO enrichment analysis of these differentially expressed genes.
Abbreviations: BP: Biological Process; CC: Cellular Component; MF: Molecular Function.
In contrast to UNDIFF, the genes up-regulated in DIFF include a different set of functional categories, namely GO: 51649: establishment of localization in cell, GO: 15631: tubulin binding, GO: 43229: intracellular organelle, GO: 8017: microtubule binding, GO: 7416: synaptogenesis, GO: 43062: extracellular structure organization, GO: 48731: system development (Table 2). This result indicates that the up-regulated genes of DIFF are involved mainly in cell differentiation, including some neuronal developments.
Differentially expressed genes in 2D and 3D differentiated hESCs
The DIFF samples were analyzed in more depth by exploring the differential gene expressions of 2D and 3D. Similar to analysis of UNDIFF vs DIFF, multiple hypothesis testing using FDR correction was used and only considered the transcripts with a greater than 2-fold difference in normalized expression between the two cell populations. Using these criteria, we identified 659 up-regulated transcripts in 2D and 381 up-regulated genes in 3D.
Of the total 1,040 genes, we found that the set of genes in 2D expressed significant annotation to GO: 51270: regulation of cell motion, GO: 44421: extracellular region part, GO: 43066: negative regulation of cell apoptosis, GO: 42641: actomyosin, GO: 45669: positive regulation of osteoblast differentiation, GO: 7507: heart development (Table 3). This indicated that the gene expression profiles of 2D are up-regulated for mesodermal lineage characteristics, although the cells were initially aimed for differentiation towards neuronal lineage.
The GO enrichment analysis of up-regulated genes in 3D revealed a different set of functional categories such as GO: 7399: nervous system development, GO: 22843: voltage-gated cation channel activity, GO: 15075: ion transmembrane transporter activity, GO: 7411: axon guidance, GO: 50767: regulation of neurogenesis (Table 3). This result indicates that the genes enriched in 3D are to support/modulate synaptic transmission, central nervous system (CNS) development, synapses formations and voltage-gated ion channel activity, indicating maturation of neurons and formation of neuronal networks.
Function 2D up |
Type |
Enrichment score |
Enrichment p-Value |
GO ID |
Extracellular region part |
CC |
11.012 |
1.65E-05 |
44421 |
Response to nutrient |
BP |
9.19531 |
0.000102 |
7584 |
Proteinaceous extracellular matrix |
CC |
7.08041 |
0.000841 |
5578 |
Extracellular matrix |
CC |
6.5108 |
0.001487 |
31012 |
Negative regulation of cellular process |
BP |
6.06385 |
0.002325 |
48523 |
Oxidoreductase activity |
MF |
5.77425 |
0.003107 |
16491 |
Regulation of muscle cell differentiation |
BP |
5.59508 |
0.003716 |
51147 |
Peptide cross-linking |
BP |
5.59508 |
0.003716 |
18149 |
Protein homodimerization activity |
MF |
5.29247 |
0.005029 |
42803 |
Function 3D up |
Type |
Enrichment Score |
Enrichment p-Value |
GO ID |
Neurotransmitter receptor activity |
MF |
9.89657 |
5.03E-05 |
30594 |
Neurotransmitter binding |
MF |
9.89657 |
5.03E-05 |
42165 |
Central nervous system development |
BP |
8.32818 |
0.000242 |
7417 |
Ion transmembrane transporter activity |
MF |
5.39393 |
0.004544 |
15075 |
Axonal fasciculation |
BP |
5.17625 |
0.005649 |
7413 |
Voltage-gated cation channel activity |
MF |
5.06092 |
0.00634 |
22843 |
Neuron migration |
BP |
4.88946 |
0.007525 |
1764 |
Positive regulation of neuron differentiation |
BP |
4.88946 |
0.007525 |
45666 |
Extracellular ligand-gated ion channel activity |
MF |
4.88946 |
0.007525 |
5230 |
Excitatory extracellular ligand-gated ion channel activity |
MF |
4.88946 |
0.007525 |
5231 |
Neuronal ion channel clustering |
BP |
4.66721 |
0.009398 |
45161 |
Neuron recognition |
BP |
4.19989 |
0.014997 |
8038 |
Regulation of axonogenesis |
BP |
3.97853 |
0.018713 |
50770 |
Regulation of neuron projection development |
BP |
3.88411 |
0.020566 |
10975 |
Voltage-gated sodium channel activity |
MF |
3.64562 |
0.026105 |
5248 |
Voltage-gated calcium channel complex |
CC |
3.3447 |
0.035271 |
5891 |
Synaptogenesis |
BP |
3.3447 |
0.035271 |
7416 |
Voltage-gated calcium channel activity |
MF |
3.20077 |
0.040731 |
5245 |
Regulation of neuron differentiation |
BP |
3.20077 |
0.040731 |
45664 |
Neuropeptide signaling pathway |
BP |
3.1572 |
0.042545 |
7218 |
Axon guidance |
BP |
3.11553 |
0.044355 |
7411 |
Regulation of neurogenesis |
BP |
3.00042 |
0.049766 |
50767 |
Synapse organization |
BP |
3.00042 |
0.049766 |
50808 |
Table 3 Differential gene expression of 2D vs 3D
Overview of 3D transcriptome
The differential gene expression level of a number of gene sets in the comparison of 3D against 2D was investigated to characterize the samples and only considered the transcripts with greater than 2-fold changes. From the heat map analyses of specific gene lists, it was observed that certain gene clusters were significantly regulated differently in 3D compared to 2D (Figure 3). For example, the well-known neuronal markers such as tubulin-β3 (TUBB3), tubulin-α 1a (TUBA1A) were increased. Common markers of neuronal cell function were also up-regulated such as neuro filament light polypeptide (NEFL), microtubule-associated protein 2 (MAP2). Neurogenic differentiation 1 (NEUROD1) is a transcriptional factor that indicates the commencement of neuronal differentiation, which was significantly increased. Also, a transcriptional factor involved in neurogenic function; delta-like 1 homolog (DLK1) was increased. Furthermore, a total of 57 transcripts of neuronal ion channel markers were examined and found that calcium channel, voltage-dependent, γ-subunit 2 (CACNG2), sodium channel, voltage-gated, type II, α-subunit (SCN2A) and sodium channel, voltage-gated, type IX, α-subunit (SCN9A) were significantly increased (Figure 3).
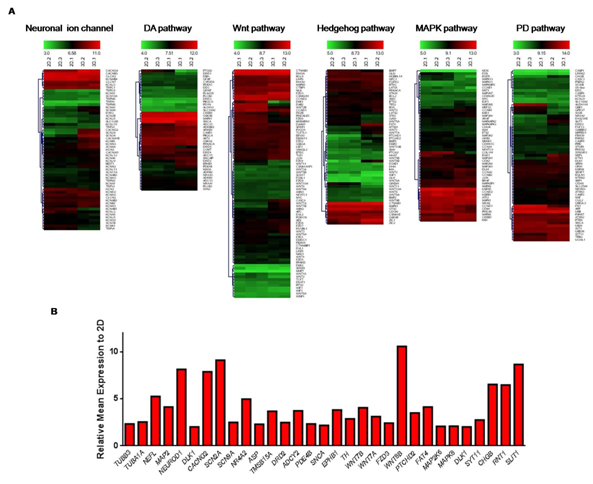
Figure 3 Differential gene expression of gene set clusters. (A) Several gene set clusters which are known to be involved in specific roles in neuronal development were investigated using heatmaps. (B) Histogram of differential genes in gene set clusters which showed at least 2-fold changes.
Next, the genes related to midbrain development were investigated which included; nuclear receptor receptor subfamily 4 group A member 2 (NR4A2), LIM homeobox transcription factor 1 α (LMX1A), LMX1B and engrailed 1 (EN1). The data indicated that no significant changes were observed except NR4A2, which increased. In addition, several genes related to SNc specific markers were examined such as insulin-like growth factor 2 mRNA-binding protein 2 (IGF2BP2), discs, large (Drosophila) homolog-associated protein 5 (DLGAP5), cyclin B2 (CCNB2), topoisomerase DNA II α 170 kDa (TOP2A), Holliday junction recognition protein (HJURP), sulfotransferase family, cytosolic, 6B, member 1 (SULT6B1), collagen, type II, α I (COL2A1), asp (abnormal spindle) homolog, microcephaly associated (Drosophila) (ASP), thymosin β 5a; thymosin β 15B (TMSB15A). None were significantly up-regulated except ASP and TMSB15A. In addition, the markers which are related to DA synthesis pathway were examined. A total of 43 transcripts were analyzed and found that DA receptor D2 (DRD2), adenylate cyclase type 2 (ADCY2), phosphodiesterase 4B, cAMP-specific (PDE4B), α-synuclein (SNCA), ephrin type-B receptor 1 (EPHB1) and tyrosine hydroxylase (TH) were increased (Figure 3).
The genes that have known or presumed to have role in regulating cell fate determination, cell polarity and DA neuronal development were examined. Wnt signaling-related genes such as wingless-type MMTV integration site family, member 7B (WNT7B), wingless-type MMTV integration site family, member 7A (WNT7A) and frizzled family receptor 3 (FZD3) were examined which showed a significant up-regulation. Of the 55 transcripts related to SHH signaling pathway, revealed that wingless-type MMTV integration site family, member 8B (WNT8B), patched domain containing 2 (PTCHD2) and protocadherin fat 4 (FAT4) were increased (Figure 3). Furthermore, 54 genes related to MAPK pathway were analyzed, showing up-regulation of mitogen-activated protein kinase kinase 6 (MAP2K6), mitogen-activated protein kinase 8 (MAPK8) and delta homolog 1 (DLK1) (Figure 3). We then examined 61 markers related to PD pathway which have been shown that dysfunction or down-regulation of these genes may lead to PD development. This gene list includes synaptotagmin-11 (SYT11), chromogranin B (CHGB), reticulon 1 (RNT1), slit homolog 1 (SLIT1) which were increased (Figure 3).
Signaling pathway analysis
The 2D vs 3D analysis that were narrowed down by only considering the transcripts greater than 2-fold changes were submitted for pathway analysis using Partek software. Seventeen pathways were found in the 381 up-regulated genes of 3D and among them, we noted “axon guidance” and “dopaminergic synapse” (Figure 4). In contrast, twenty-five pathways were significantly found in the up-regulated transcripts set of 2D and among them, we noted “focal adhesion” and “Transforming growth factor (TGF)-β signaling pathway” (Figure 5). Out of these pathways, the top six pathways from both of samples were noted and the two pathways were examined in details; axon guidance and focal adhesion pathways.
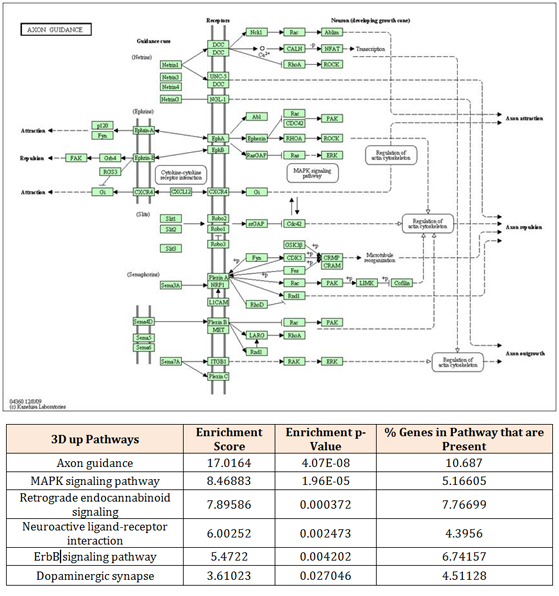
Figure 4 Significant pathway in 3D. The 381 up-regulated mRNA set in 3D are significantly enriched by mRNA coding for proteins involved in axon guidance pathway.
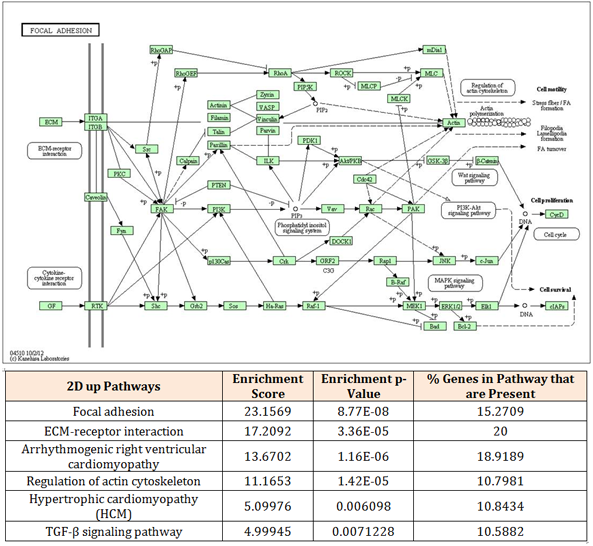
Figure 5 Significant pathways in 2D. The 659 up-regulated mRNA set in 2D are significantly enriched by mRNA coding for proteins involved in focal adhesion pathway. Both pathway pictures were imported from KEGG pathway website. The biological relationships between two proteins are represented as a line (an edge).
The axon guidance pathway contained 14 genes representing 10.69% of the genes detected by KEGG pathway analysis. The axon guidance pathway is dominated by genes regulating axon formation such as deleted in colon cancer (DCC), unc-5 homolog A (UNC5A) and ephrin type-A receptor 7 (EPHA7). On the other hand, the focal adhesion pathway contained 20 genes representing 15.27% of the genes detected by KEGG pathway analysis. It is dominated by genes associated with extracellular matrix, promoting cell adhesion and cell differentiation such as reelin (RELN), fibronetin (FN1) and vitronectin (VTN).
The data generated from the pathway analysis function using Partek software revealed that the signaling molecules in 3D showed high enrichment of MAPK, retrograde endocannabinoid and ErbB signaling pathways, which are involved in neuronal differentiation. For example, retrograde endocannabinoid pathway is involved in regulating/maintaining synapse-specific refinement in various regions of brain.8 On the other hand, 2D showed significant enrichment of gene clusters in the TGF-β pathway. TGF-β pathway has been reported to regulate cell growth, cell differentiation, particularly osteogenesis and apoptosis.9
Validation of microarray data by qPCR
To validate the results from the microarray assays, qPCR was performed with the samples used for the microarray analysis with an additional sample of E2 cell line, passage 36 which has been differentiated under 3D platform with PA6 cells for 28 days (resulting total of 3 biological replicates for each group; E2, 2D and 3D). Out of the genes which were previously investigated, a total of 10 genes (2 genes from each gene set tested) were selected which showed up-regulation in 3D samples; SYT1, SLIT1, CACNG2, SCN2A, MTSS1, FAT4, DRD2, SNCA, EPHB1, MAPK8. Using the ΔΔCt method to determine fold differences of relative gene expression in 2D vs 3D, the qPCR experiments largely confirmed the results from the microarrays (Figure 6). Although we observed variability between the samples, there was an overall consistency between both methodologies demonstrating a broad up-regulation of the selected genes. However, CACNG2, SCN2A and DRD2 did not reach significance in the qPCR results using Student’s t-test, unpaired, two-tailed statistics method.
The microarray data presented in this study were compared for the gene expression profiles of undifferentiated and differentiated hESC samples under conventional 2D microenvironment or novel 3D system via microencapsulation. As expected, the global gene expression analysis of UNDIFF vs DIFF showed significant down-regulation of currently recognized pluripotent stem cell markers such as POU5F1, TDGF1, NANOG, UTF1 and DNMT3B in DIFF. In addition to this, GO enrichment analysis of UNDIFF showed high enrichment of gene ontologies related to cell proliferation or mitosis whereas differentiation-related gene ontologies were highly enriched in DIFF. The down-regulation of tumorigenic genes such as POU5F1, NANOG and UTF1 in DIFF may provide evidence for the non-tumorignic nature of the examined DIFF and their safety for potential use in cell-based therapy for PD downstream. However, in vivo analysis by transplanting the DIFF samples into SCID mice is required.
The study has investigated the neuronal expression profiles of DIFF samples which included a novel 3D-derivation of DA neurons, of which none have yet been published. The data showed that genes which are involved in development of microtubules such as TUBB3 and transcription factors which commence the neuronal differentiation such as NEUROD1 were significantly up-regulated in 3D. Furthermore, several neuronal ion channel markers such as CACNG2, SCN2A and SCN9A were highly enriched in 3D. It is expected that the hypoxic environment within the embryoid bodies (EBs) like in 3D cultures may have increased the rate of DA neuronal expressions which has been previously reported that the cells in hypoxic culture condition enhance DA neurogenesis.10 In addition, it has been shown that cells cultured in a 3D environment can freely interact with neighbouring cells and respond more readily to each other, which significantly affects stem cell fate decisions such as apoptosis, self-renewal and differentiation.11,12 In comparison to the cell-cell interaction and the hypoxic environment of EBs, the conventional 2D differentiation system has limited cell-cell contact with its neighbour cells and higher exposure to oxygen may have led to slower DA neuronal expressions than differentiated ES cells under 3D environment. Concurrently, GO enrichment analysis indicated that the differentially expressed gene ontologies of the 3D showed higher enrichment of neuronal development whereas 2D was enriched in mesodermal differentiation-related gene ontologies. The data corresponds to the electrophysiology data (unpublished) which indicated potential Na+ and K+ fluxes and a graded potential in 3D differentiated hESCs whereas no responses were observed in 2D differentiated cells. The neuronal gene expression profile analysis also suggests that the 3D requires further up-regulations of the other ion channel subunits e.g. voltage-gated potassium channel subunit β -1 (KCNAB1), potassium channel subfamily K member 1 (KCNK1) and potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 1 (HCN1) to be mature electrophysiologically functional neurons.
In many published studies, the expression of only a single TH or 2-3 markers for DA neurons e.g. aromatic l-amino acid decarboxylase (AADC), dopamine transporter (DAT), vesicular monoamine transporter 2 (VMAT2) have been used to indicate dopaminergic identity of neurons. The markers which are related to midbrain and DA synthesis showed significant increase in 3D when compared to 2D such of NR4A2, TH, DRD2, ADCY2, PDE4B, SNCA and EPHB1. However, other midbrain-related transcripts such as LMX1A, LMX1B and EN1 were not significantly increased in 3D. In addition, SNc specific markers such as IGF2BP2, DLGAP5, CCNB2, TOP2A, HJURP, SULT6B1 and COL2A1 showed no significant up-regulations. This may suggest that the cells in 3D have differentiated to early DA neurons at a faster rate than 2D and are yet to fully express the midbrain relevant markers.
One focus of our study was to characterize a complex series of signaling programs potentially involved in the priming process of the 3D samples. A better understanding of these signaling pathways at a cellular level would be of great assistance to the elucidation of the mechanisms underlying stem cell lineage specification and to the discovery of potential targets for stem cell therapy, gene therapy and pharmaceuticals. The comparison of gene expression profiling in signaling-related genes of the 3D and 2D samples was carried out by importing the gene lists obtained from PCR assays from SA Biosciences. It was noticed that some known and required pathways for induction and formation of DA neurons were activated in 3D, including Wnt, SHH, MAPK and PD pathways. Wnt signalling pathway has been reported widely to control diverse processes such as cell proliferation, self-renewal, cell polarity, cell death and cell fate specification.13 The data has shown significant up-regulation of Wnt signaling genes in 3D such as WNT7B, WNT7A and FZD3. WNT7B and WNT7A have been shown to be involved in several developmental processes, including regulation of cell fate and patterning during embryogenesis.13-15 The gene FZD3 has previously been reported that mutant mice with defects in frizzled family receptors such as FZD3 showed a severe impairment of midbrain morphogenesis.13,16 However, not many genes related to Wnt signaling pathway were significantly up-regulated to draw out a definite conclusion. The genes related to SHH signaling pathway which has been reported to be involved in directing neural progenitors (NPs) to acquire specific cell identifies and induction of cell types of the telencephalon, diencephalon, midbrain and anterior hindbrain were also examined. The signaling-related genes such as WNT8B, PTCHD2 and FAT4 were up-regulated in the 3D samples. WNT8B and PTCHD2 have been shown to be involved in the developing brain, whereas FAT4 is involved in planar cell polarity.17-20 MAPK signalling pathway has been previously reported to be involved in neuronal functions connected to synaptic plasticity. Among the 54 genes which are related to MAPK pathway, few genes showed significant up-regulation such as MAP2K6, MAPK8 and DLK1. MAPK2K6 and MAPK8 have been known to be one of the cascade activators in MAPK pathway which are involved in cellular processes such as stress-induced cell cycle arrest, transcription activation and apoptosis.21,22 The gene DLK1 has been shown to inhibit adipogenesis.23 In addition, the PD-related gene expressions were analysed which have been shown that dysfunction or down-regulation of the genes may lead to PD development.24 Several genes showed higher expression in 3D such as SYT11, SLIT1 and RNT1 which suggests that; (i) the 3D system enabled faster rate of DA neuron derivation and (ii) neurons which are more mature than 2D-derived neurons which can secrete higher DA level under electrophysiological changes. The gene SYT11 is involved in maintenance of synaptic function and represents a component of Lewy bodies in brains of PD patients.25 Using SLIT1 knockout mice, it was indicated that SLIT1 is critical during the initial establishment of dopaminergic pathways, with roles in the dorso-ventral positioning and precise pathfinding of these ascending longitudinal axons.26 In addition, RTN1 gene has been suggested that the dysfunction of RTN1 gene leads to aberrant protein folding and aggregation in endoplasmic reticulum (ER). This induces ER stress and deficits in ER chaperone function, which may lead to PD.27,28 Although there are some suggestive data that some of the signalling transcripts which direct midbrain DA neuronal differentiation are up-regulated in the 3D samples, there were several genes-related to signalling pathways that were not significantly changed and remain to be further investigated.
In the present study, KEGG pathway analysis of the up-regulated genes of 2D and 3D samples was conducted using Partek software. Six molecular pathways were significantly enriched in 2D samples: focal adhesion, extracellular matrix (ECM)-receptor interaction, arrhythmogenic right ventricular cardiomyopathy (ARVC), regulation of actin cytoskeleton, hypertrophic cardiomyopathy and TGF-β signaling pathway. The cell cycle pathways in 2D are dominated with gene sets which are involved in regulating mesodermal differentiation. It has been reported that ectodermal differentiation inhibits the mesodermal differentiation mediated by TGF-β signaling pathway.9,29,30 Therefore, it may be interpreted that the cells in 2D are still in the transitional stage between ectodermal and mesodermal differentiation where the TGF-β signaling molecules are still active. For 3D, six molecular pathways were significantly enriched; axon guidance, cell adhesion molecules, MAPK pathway, neuroactive ligand-receptor interaction, ErbB signaling pathway and dopamineric synapse. The cell cycle pathways in 3D are dominated with genes regulating neuronal and DA differentiation. This suggests that the TGF-β signaling genes in 3D may have been inhibited, allowing the transition of mesodermal to ectodermal differentiation. In addition, the significant pathway analysis corresponds to the analysis of individual signalling transcripts of Wnt, SHH and MAPK pathways, suggesting the 3D samples have differentiated further towards neuroectodermal/dopaminergic lineage than 2D samples.
In conclusion, the experiments herein suggested that the transcriptome of hESCs differentiated under 3D platform may have enhanced the DA neuronal differentiation rate. Although further dissection of signalling pathways involved in promoting dopaminergic formation is needed, our results suggest that pathways which are related to DA neuronal differentiation are activated. In addition, further differentiation period of both 3D and 2D systems in the future would provide valuable information in understanding the molecular interactions involved in the DA neuronal development.
This study was funded by The NHMRC Program Grant (PSS) and A Special Stem cells Grant by The Faculty of Medicine, University of New South Wales, Australia (PSS, KS)' and the publication is supported by the Department of Neurology, Hianjin Huanhu Hospital, China (PZ).
The author declares no conflict of interest.

©2017 Kim, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.