Journal of
eISSN: 2475-5540


Bone marrow derived stem cells could offer significant multipotent differentiation capacity and plasticity in other species. However, in rat marrow there are few reports. This study described, simple and easy method for isolation of rat bone marrow stem cells (rBMSC) based on their density and plastic adherence properties and along with their trilineage differentiation potential. Rat bone marrow stem cells were isolated by density gradient and plastic adherence method. After isolation of stem cell they were characterized by phase contrast microscopy and flow cytometry. Moreover, differentiation into osteoblast, adipocytes and chondrocytes of mesenchymal stem cells (MSC) on different induction medium was analyzed using RT-PCR and their respective staining methodology. The attached nucleated cells showed their spindle shaped fibroblastic like morphology. Flow cytometry results revealed that they were negative for haematopoietic markers such as CD 31 and CD 45 and positive for mesenchymal markers CD 90 and CD 29. On the other hand, they were differentiated into osteogenic, adipogenic and chondrogenic and this was confirmed by alizarin red, Oil red O and Alcian blue stain respectively. Further RT-PCR results indicated that rBMC expressed Runx2; Ppar-γ and Aggrecan genes which showed that these cells can easily differentiate into many different lineages and this can be applicable for cell based regenerative disorders.
Keywords: rat bone marrow, mesenchymal stem cell, cd markers, osteogenic, adipogenic, chondrogenic
rBM-MSC, rat bone marrow derived mesenchymal stem cells; CFU-F, colony forming unit-fibroblast; DPBS, dulbecco’s phosphate buffer saline; DMEM, dulbecco’s modified eagles medium; RT PCR, reverse transcription polymerase chain reaction; RNA, ribonucleic acid
Bone marrow hosts variety of tissues including hematopoietic lineage cells and mesenchymal stem cells (MSCs). The hematopoietic cells are the major source of the blood cells in the adult body which are regulated within the microenvironment of the stromal cells of the bone marrow.1,2 MSCs are multipotent cells and can be differentiate into various cell lineage depend upon their environment and culture condition in which they are kept. Rat bone marrow derived stem cells (rBM-MSCs) have long term self renewing and capabilities of pluripotency including osteoblast, adipocytes, chondrocytes, tenocytes, muscle cells and neurogenic cells, which make them ideal source of stem cells for regeneration of injured tissue.3–5
Bone marrow derived mesenchymal stem cells have been isolated from different species. Report says that each species having different culturing and growing capacity.6 For example human bone marrow derived stem cells are easy to harvest and maintain in culture condition where as rat MSCs is more difficult compared to other species.7,8 Technical difficulties in isolation limited the number of animal experiment and its required animal transplantation model for pre clinical studies.9 Density gradient centrifugation,10 plastic adherence11 and immunomagnetic selection12,13 like various methods are available for isolation of MSCs from bone marrow. No appropriate methods are available for selection of suitable cells. Each method has their own pros and cons. Therefore this study combined density gradient centrifugation and plastic adherence for easy and reliable method for selection of suitable cells.
Collection of bone marrow
Four male wistar rats aged 8-14weeks old were prepared for collection of bone marrow. All procedure was undertaken after getting prior approval from Institutional Animal Ethics Committee (IAEC). Briefly after euthanasia, animals back side were cleanly shaved and prepared aseptically. The skin incision was made on the lateral aspect of thigh and both femur and tibia from one rat were taken after stripping out adherent muscles of the knee end. The collected bone was taken to laminar hood and placed in Petri dish containing DMEM (Dulbecco’s Modified Eagles Medium) media with antibiotics. Both the metaphyseal region of femur was cut with scissors and needle was inserted into the bone to aspirate the content by flushing with 1ml of complete media. The cell suspension was centrifuged at 980rpm for 5min to concentrate the cells. The cell pellets were resuspended with 5ml of complete DMEM medium then layered over HISTOPAQUE -1077(Sigma) and centrifuged at 2500rpm for 30minutes. Mononuclear cells were collected from the interface by gradient centrifugation and washed with Ca+ and Mg+ free Dulbecco’s Phosphate Buffer Saline (DPBS). The cell count and viability was done after addition of Trepan blue dye in the automatic cell counter then it was seeded in 1x106/ml concentration in a 25cm2 flask. Non- adherent cells were removed after 72hrs of cell seeding. Media was added every 3 or 4days once until attaining confluence.
Culture conditions
The adherent cells were supplemented with complete medium containing DMEM, 15% fetal bovine serum (FBS-Gibco) and 1% antibiotic mixture of 100units/ml of penicillin and 100µg/ml of streptomycin (Invitrogen/Gibco). The cells were maintained in a humidified chamber at 37°C which supplemented with 5% CO2. After attaining 70-80% confluence, culture was passage with Trypsin-EDTA solution (0.25% Trypsin and 1Mm EDTA (Gibco)). Third passage cells were used for further differentiation studies.14
Colony forming assays
For this assays, 2 cells per cm2 were plated in 100mm tissue culture dish and maintained up to 14 days with complete media @ 37°C in 5% humidified CO2. At the end of assay, cells were washed with DPBS and fixed in methanol for 5 minutes. Later, the cells were stained with Giemsa for 30 minutes at room temperature. Then the plates were washed with distilled water, dried and cells were counted. Those, faintly stained cells were ignored.15
Cell viability assay
The 3-(4, 5-Dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide (MTT) (Sigma) assay was used to detect the number of proliferating cells in culture based on the reduction of MTT by mitochondrial dehydrogenases. Third passage cells were plated in 96 well plate at 1x104 cells/well in complete culture media for 1, 3, 7, and 14 days of incubation. Following incubation, the cells were treated with 20µl MTT (5mg/ml) for 4hrs at 37°C in a 5% CO2 incubator. After that media was removed gently without disturbing attached monolayer and dried for few minutes. 150µl of DMSO was added to all the wells and mixed gently to dissolve the fromazan crystals without forming air bubbles. The absorbance level was measured at 570nm wavelength in ELISA reader. The experiment was conducted in triplicate and DMSO as the blank.
Flow cytometric analysis of CD markers
Flow cytometric was performed in FACS caliber (BD, Frankin lakes, NJ, USA). The cells were centrifuged at 1200rpm for 5min after trypsinization then pellet was dissolved with PBS at the concentration of 1x106/ml. The cells were incubated with primary polyclonal anti rat antibody against positive markers and negative markers likely CD-29, CD-90 and CD-31, CD-45 respectively at room temperature for 40 minutes. After the cells were washed twice with PBS containing 2% bovine serum albumin and centrifuged at 1500rpm for 5min, they were resuspended with FITC labeled secondary antibody for 30 minutes at 4°C. Cell fluorescence was evaluated in FACS caliber and data were analyzed using cell quest software (Beckon Dickinson). The experiment was conducted in triplicates.
Osteogenic differentiation
Third passage cells were counted and seeded on 24 well tissue culture plate at 2x106 cells per well. After attaining 80% confluence, four wells supplemented with osteogenic differentiation medium (n=4) (Stem pro Gibco) where as other wells supplemented with complete medium served as negative control. Media was changed weekly twice and maintained up to 21 days. The differentiation potential of osteogenesis was evaluated by alizarin red and alkaline phosphatase staining for calcium deposition and osteogenic specific marker RunX-2 was analyzed through RT-PCR.
Adipogenic differentiation
To induce adipogenic differentiation, third passage cells were seeded on to 24 well plate at 2x106 cells/ well (n=4). Cells were replenished with adipogenic differentiation media (Stem pro/Gibco) after reaching 80% confluence. Culture was maintained up to 3 weeks and media changed twice a week. Presence of Lipid droplets in the cells indicated that positive for adipogenic differentiation which was confirmed by Oil Red O (Sigma) staining with 0.5% oil red O in methanol. Expression of adiopogenic marker Ppar-γ was confirmed by RT-PCR.
Chondrogenic differentiation
The chondrogenic differentiation was assessed by using stem pro chondrogenic induction medium (Stem pro/Gibco). Culture condition was same as mentioned in osteogenic differentiation (n=4). Culture was maintained up to 18 days. Alcian blue staining was used to assess the chondrogenic differentiation and specific marker Aggrecan was confirmed through RT PCR.
Reverse transcription polymerase chain reaction (RT PCR)
RT-PCR analysis was done to detect specific differentiation potential of rat MSC cells on standard protocol. Total RNA was isolated from differentiated rat bone marrow mesenchymal stem cells using RNeasy mini kit (Qiagen, USA) according to the manufacture protocols. To obtain pure RNA, the RNA was treated with DNase which preventing the DNA carryover in the final preparation of RNA. Total concentration of RNA was measured by Nano drop 1000 spectrophotometer and measured in nanogram per micro litter. The first strand cDNA was synthesized from total RNA (1µg) using Murine Leukemia Virus reverse transcriptase (Fermentas ) for 30 minutes at 42°C in the presence of oligo- dt primer. The PCR was carried out with cDNA template of following gene of interest RunX-2, Ppar-γ and Aggrecan. Oligonucleotide primers and their temperature conditions are given in Table 1.
Name |
Sequence |
Product size (bp) |
Annealing temperature (°C) |
RunX2 |
Forward 5'- GCCGGGAATGATGAGAACTA-3' |
155 |
52 |
Reverse 5'- TTGGGGAGGATTTGTGAAGA-3' |
|||
PPARγ |
Forward 5'- CGTCCCCGCCTTATTATTCTGA-3' |
456 |
54 |
Reverse 5'- CCTCGCCTTGGCTTTGGTC-3 |
|||
Aggrecan |
Forward 5'-CAAGAATCAAGTGGAGCCGTGTT-3' |
295 |
54 |
Reverse 5'-TCAAAGTCCAGTGTGTAGCGTGT-3' |
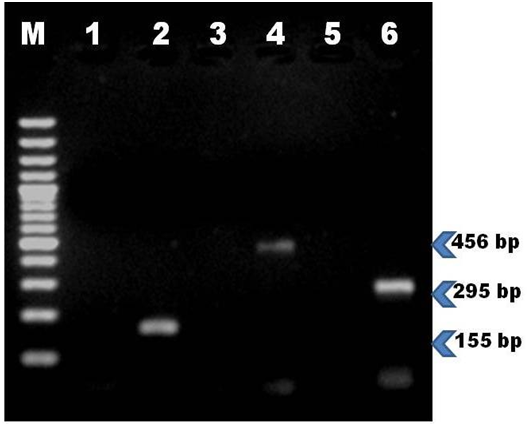
Table 1 Gene specific primers used in the reverse transcription polymerase chain reaction (RT-PCR)
Statistical analysis
All data were presented as Mean±SE. One way ANOVA and tukey post- hoc test were performed to compare the groups. p<0.05 was considered to be statistically significant.
Cell culture of rBMSC
The culture was observed by using an inverted microscope. On initial seeding, the heterogeneous population of cells was round oval in shape and floating over the media. MSC cells were started to attach sparsely in culture flask and other non adherent hematopoietic cells were eliminated from culture after subsequent media change. Cells started to grow in smaller colonies after 3 or 4th day of culture (Figure 1A). As growth continued cells started to change their morphology from round cells to spindle shape which morphologically defined as fibroblast like cells (Figure 1B). This fibroblast like cells started to interconnect with other cells and monolayer confluence was attained after 15-17days of culture (Figure 1C). During the passaging, the morphology of cells changed gradually and exhibit more flattened fibroblast like colonies.
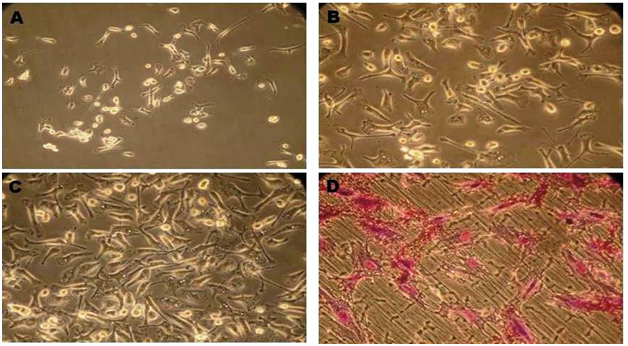
CFU- assay
Colony forming assay suitable assay for assessing proliferating and colonies forming capacity of cells on third passage was measured. When cells plated at low density it was forming colonies (Figure 1D).
MTT assay
Under the culture condition the proliferative capacity of rBMSC was assessed by MTT test. Cell livability / proliferation was significantly (p<0.05) increased from day 1 to 14 (n=3) Mean±SE 0.22±0.02, 0.28 ± 0.02, 0.51±0.08 and 0.61±0.05 respectively (Figure 2).
Characterization
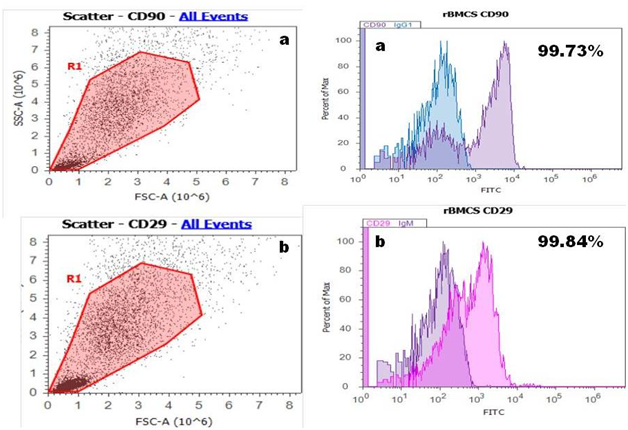
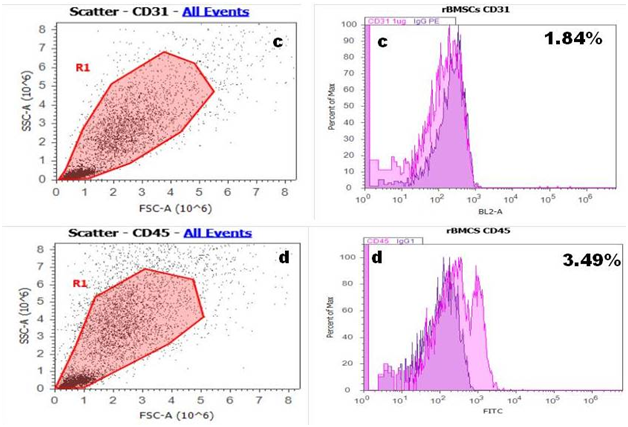
Flow cytometry: Majority of rBMSC expressed the surface marker positive for CD 90 and CD 29 Figure 3A & 3B and relatively negative for hematopoietic stem cell marker CD 31 and CD 45 these results shown in Figure 3C & 3D which indicate that most of the cells derived from bone marrow were mesenchymal.
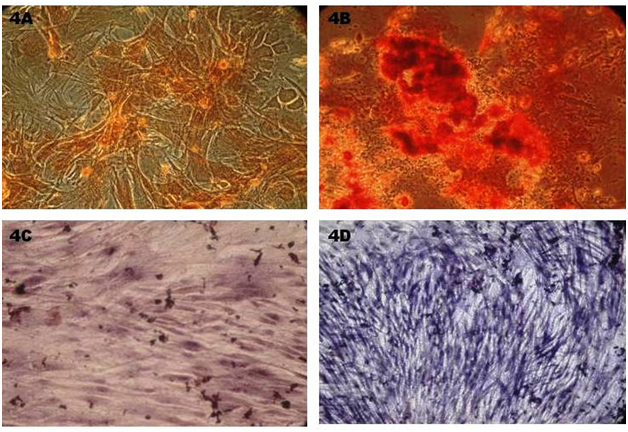
Osteogenic differentiation: The rMSC cells differentiated into osteogenic lineage after 18 days of culture in osteogenic induction medium which was confirmed by presence of extracellular mineralization as evident on alizarin red staining and alkaline phosphatase staining. The positive osteogenic culture showed more intense alizarin red staining, indicating that more calcium deposition. Control culture showed least background staining (Figure 4A & 4B). Alkaline phosphates staining intensity was evident in differentiated monolayer rBM-MSC compare to control cells (Figure 4C & 4D). Further osteogenic specific gene marker Runx2 was confirmed through RT-PCR which was shown by 155bp.
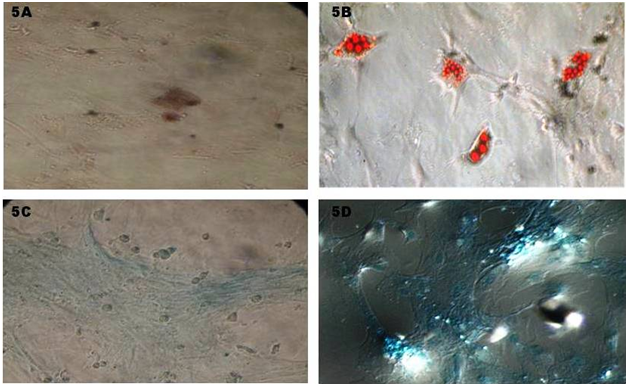
Adipogenic differentiation: Adipocytes differentiation from rat MSC was observed after 7 days of in vitro culturing which supplemented with adipocytes induction medium. Cells were ability to change their morphology from spindle shape to flat one. There was a discrete lipid droplet in cytoplasm of the cells which confirmed by oil red O staining (Figure 5A & 5B). This shows rMSC cell can differentiate in to adipocytes cells. Supplementary to above, specific gene for adipocytes (Ppar-γ) expression was detected by RT-PCR.
Chondrogenic differentiation: Cells from third passage were subjected to chondrogenic differentiation and after attaining confluence cells were supplemented with chondrogenic induction medium. Cells started to change their morphology (polyclonal and round) in time dependant manner compare to control which was showed on Alcian blue staining (Figure 5C & 5D). Further this was confirmed with RT-PCR for Aggrecan specific marker for chondrocytes (Figure 6).
Among the various stem cells, Bone marrows derived MSC cells could be used for regenerative medicine not only due to their self renewing capacity and differentiate into various functional cell types in specific tissue but also due to their homing capacity and non immunogenic nature.16–18 Inconsistent results were obtained by various authors during Isolation and defining the characteristic of MSC, as because isolation of MSC varied dramatically due to their species difference and adopting methodologies for expansion and plating of cells.19,20 However bone marrow stem cells can be isolated by Ficoll gradient centrifugation, plastic adherence. In Ficoll gradient centrifugation cells were selected based on the relative density of the cells. Due to plastic adherence nature of stem cells they can be obtained easily but getting of pure stromal cells are difficult. Other methods could be used to isolate stem cells but none of these methods were found suitable.21,22 This study effectively used both Ficoll gradient centrifugation and plastic adherence methodologies for BMSC isolation.
Initially adhered MSC cells were expanded into round cells then it slowly become spindle shaped morphology and forming colonies of fibroblast when plated in the low density. These results are consistent with various other studies.23,24 Further, in this study was to investigate cell surface markers. According to the international society for cellular therapy (ISCT) MSC should posses some CD markers to define stem cells. To prove this concept, performed Flow cytometric analysis for MSC which showed that isolated bone marrow stem cells were positive for CD 90 and CD 29 and lack of expression of CD 31 and CD 45 indicating that they were from non haematopoietic lineage and these results were reliable with previous reports.25,26
Colony forming unit - fibroblast (CFU-F) assay was used to measure the cell proliferation and Colonogenic capacity. The results showed that rMSC ability to proliferate and form colonies of fibroblast in particular intervals.27 Trilineage capability of rMSC was assessed by keeping cells in different induction medium compare to control media. Isolated stromal cells were ability to differentiate into other mesenchymal cell lineages like osteoblast, adipocytes and chondrocytes as observed by other workers.28–30 Osteogenic, adipogenic and chondrogenic differentiation potential was confirmed with special staining viz, Alizarin red, Oil Red O, Alcian blue respectively. In addition, these results were further confirmed with expression of specific gene like Runx2, Ppar-γ and Aggrecan for osteogenic, adipogenic and chondrogenic respectively.
In summary, the combination of density gradient centrifugation and plastic adherence is an efficient method for producing a homogenous population of bone marrow stromal cells. The data further demonstrated that MSC has ability to differentiate into highly specialized cells and each with specialized cell function. This pluripotency and self renewing capacity make them ideal source for regenerative medicine.
None.
The author declares no conflict of interest.

© . This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.