Journal of
eISSN: 2475-5540


Mini Review Volume 1 Issue 1
Department of Genetics, Cell Biology and Developmental Biology, University of Minnesota, USA
Correspondence: Nobuaki Kikyo, Stem Cell Institute, Department of Genetics, Cell Biology and Developmental Biology, University of Minnesota, USA, Tel 612 624 0498, Fax 612 624 2436
Received: June 10, 2015 | Published: November 27, 2015
Citation: Lowe M, Hostager R, Kikyo N. Preservation of epigenetic memory during DNA replication. J Stem Cell Res Ther. 2015;1(1):39-42. DOI: 10.15406/jsrt.2015.01.00007
Faithful duplication of a cell’s epigenetic state during DNA replication is essential for the maintenance of a cell’s lineage. One of the key mechanisms is the recruitment of several critical chromatin modifying enzymes to the replication fork by proliferating cell nuclear antigen (PCNA). Another mechanism is mediated by the dual function of some histone modifying enzymes as both “reader” and “writer” of the same modification. This capacity allows for parental histones to act as a seed to copy the modification onto nearby newly synthesized histones. In contrast to the vast quantity of research into the maintenance of epigenetic memory, little is known about how the recruitment of these maintenance enzymes changes during stem cell differentiation. This question is especially pertinent due to the recent emphasis on cell reprogramming for regenerative medicine.
Keywords: chromatin, DNA replication, DNA methylation, epigenetics, histone modification
ASF1, anti-silencing factor 1; CAF-1, chromatin assembly factor 1; DNMT1, dna (cytosine-5-)-methyl transferase 1; EED, embryonic ectoderm development; EZH2, enhancer of zeste homologue 2; FACT, facilitates chromatin transcription; HAT1, histone acetyl transferase 1; HDAC1, histone deacetylase 1; HP1, heterochromatin protein 1; iPS cell , induced pluripotent stem cell; MBD1, methyl-cpg binding protein 1; MCM2-7, mini-chromosome maintenance 2-7; MLL1, mixed lineage leukemia 1; NAP1, nucleosome assembly protein 1; NP95, nuclear protein 95; ORC, origin recognition complex; PCNA, proliferating cell nuclear antigen; PIP-Box, pcna-interacting protein-box; PHD, plant homeo domain; PRC2, polycomb repressive complex 2; SMARCA5, swi-snf related, matrix-associated, actin-dependent regulator of chromatin, subfamily a, member 5; Suv39H1, suppressor of variegation 3-9 homolog 1; WSTF, williams syndrome transcription factor
The epigenetic state of a given cell must be accurately duplicated on daughter chromosomes during DNA replication to maintain the identity of the cell. Because epigenetic modifications are conferred by modification-specific enzymes, a central question is how appropriate epigenetic enzymes are recruited to newly assembled daughter chromatin in a locus-specific manner. Due to human DNA being replicated at a speed of 2 to 3kb per min1 and the replication fork associating with more than 400 chromatin factors,2 there must be a robust system in place to accurately duplicate the chromatin structure during replication. Among the mechanisms involved in the preservation of epigenetic memory during DNA replication, this review article will only focus on the two most extensively studied groups of proteins as the primary recruiters for epigenetic enzymes: DNA replication factors and preexisting modifications on parental histones. Epigenetic inheritance during DNA replication is a vast field and more comprehensive reviews are available in references.3–5
Recruitment of epigenetic enzymes by DNA replication factors
Proliferating cell nuclear antigen (PCNA) forms a homo trimeric ring surrounding the parent DNA duplex and serves two functions during DNA replication.6 First, it forms a sliding clamp that tethers DNA polymerase δ (for the lagging-strand DNA) and ε (for the leading-strand DNA) onto template DNA. This tethering prevents the dissociation of the polymerases from DNA and leads to increased processivity of the polymerases. Second, PCNA recruits more than 50 different proteins that contain a conserved sequence called the PCNA-interacting protein-box (PIP-box) with a few exceptions.7 The interacting proteins include chromatin modifying enzymes (Figure 1A), in addition to those involved in DNA replication, DNA repair, cell cycle control, transcription, and chromatin assembly.6,7
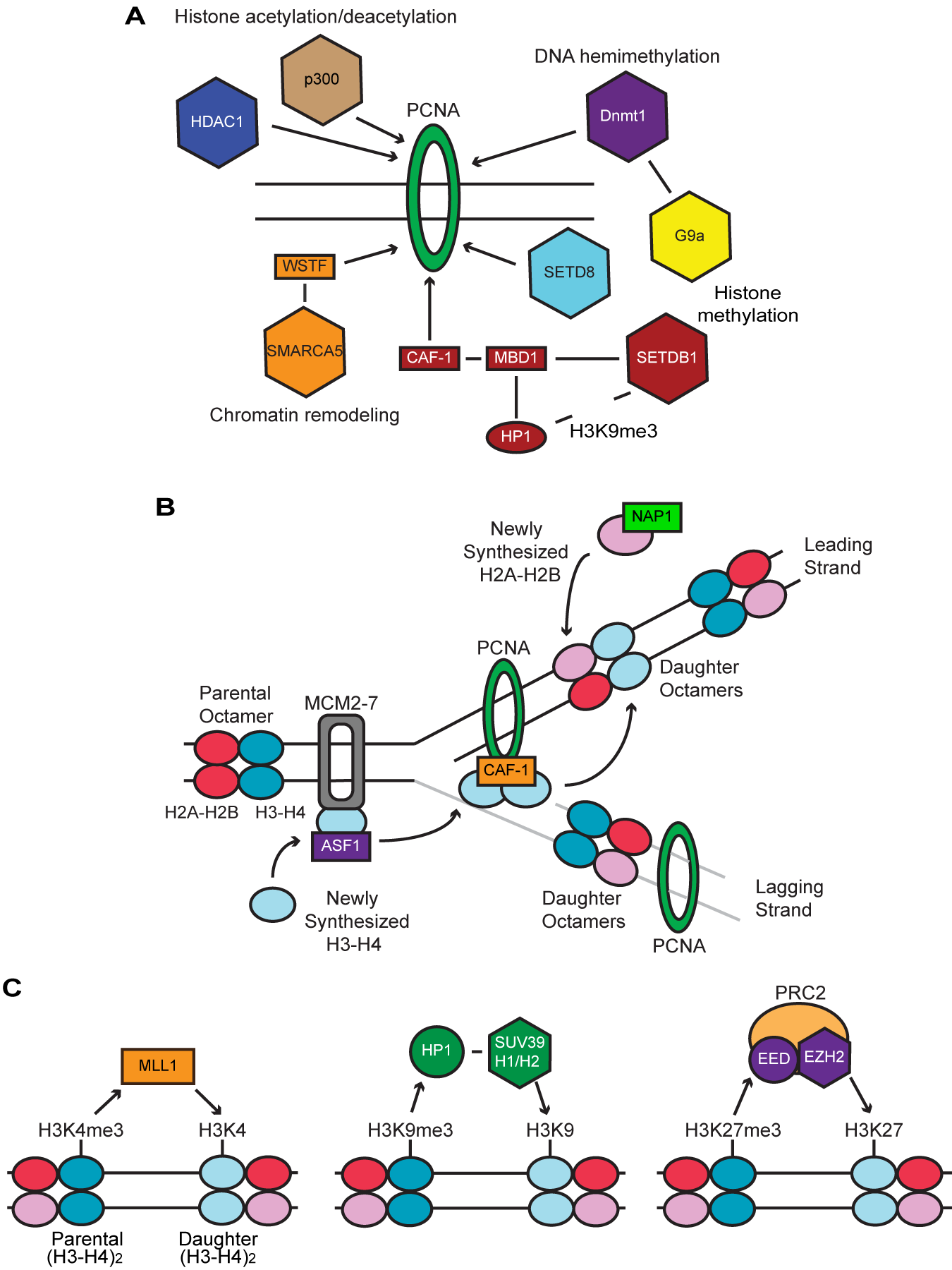
Figure 1A Chromatin modifying enzymes bound by PCNA while on the DNA duplex.
PCNA recruits these chromatin modifying enzymes directly or via an intermediate protein. Four main sets of modifying enzymes are shown, each with a unique effect on chromatin.
Figure 1B Assembly of histone octamers onto the daughter strand at the DNA replication fork.
The parental octamers dissociate into two H2A-H2B dimers and a single (H3-H4)2 tetramer, which are then transported across the replication fork and deposited onto the daughter duplexes. This distribution is random and thus the daughter octamers are likely to contain both a newly synthesized and parental H2A-H2B dimer as well as either a newly synthesized or parental (H3-H4)2 tetramer as displayed in the diagram. The newly synthesized H3-H4 dimers are carried across the replication fork by ASF1 and CAF-1 prior to forming a tetramer and deposition onto the daughter DNA duplex. The newly synthesized H2A-H2B dimers are likewise deposited onto the daughter duplex via NAP1.
Figure 1C Duplication of histone modifications from parental to daughter octamers via enzymes or complexes with both a recognition and catalytic capacity. These enzymes recognize a modification present on the parental (H3-H4)2 tetramer and then mediate the same modification onto the daughter tetramer, allowing for propagation of the modification.
Among the chromatin modifying proteins, DNA (cytosine-5-)-Methyl Transferase 1 (Dnmt1) is a prime example for the preservation of epigenetic information during DNA replication through a direct or indirect interaction with PCNA.8 Dnmt1 induces methylation of CpG sites on the newly synthesized DNA strand by duplicating the methylation pattern of the parental DNA strand (hemimethyltransferse) and thus functions as a maintenance methyl transferase. As for histone modifying enzymes, PCNA recruits the acetyl transferase p300,9 the methyl transferase SETD810 and histone deacetylase 1 (HDAC1).11 In addition, PCNA indirectly recruits via Dnmt1 the histone methyl transferase G9a, which mediates the dimethylation of lysine 9 on histone H3 (H3K9me2), a typical marker for heterochromatin.12 Furthermore, PCNA uses the histone chaperon chromatin assembly factor 1 (CAF-1)13 as an indirect recruiter for methyl-CpG binding domain protein 1 (MBD1), which then interacts with heterochromatin protein 1 (HP1)14 and the histone methyl transferase SET domain bifurcated 1 (SETDB1).15 SETDB1 mediates H3K9me3, to which HP1 binds, maintaining the heterochromatin structure.16 As an example of chromatin remodeling ATPases, Williams syndrome transcription factor (WSTF) forms a protein complex with the chromatin remodeling ATPase SMARCA5 (SWI-SNF related, matrix-associated, actin-dependent regulator of chromatin, subfamily a, member 5, also called SNF2H) and increases chromatin accessibility.17 In addition to PCNA, the origin recognition complex (ORC) and DNA polymerases are known to maintain epigenetics in yeast and Drosophila but their roles in mammalian cells are far less clear than those of PCNA.18
While these molecular interactions explain how a handful of key chromatin modifying enzymes are recruited to the replication fork, it remains largely unknown how PCNA and other replication proteins replace their recruitment partners in a locus-specific manner while the replication fork is rapidly moving along the parent DNA duplex. The primary cue for the locus-specific recruitment is thought to be provided by the recycled parental histones on the daughter DNA duplex as described next.
Recruitment of epigenetic enzymes by re-deposited parental histones
Nucleosome disassembly and reassembly during DNA replication have been thoroughly reviewed in references.19–21 The parental DNA duplex is unwound during replication by the replicative DNA helicase complex mini-chromosome maintenance 2-7 (MCM2-7). The interaction between histone octamers and the DNA duplex is destabilized during this process. The parental histone octamers are dissociated into two H2A-H2B dimers and one (H3-H4)2 tetramer, which is then randomly distributed onto the two daughter DNA duplexes. This means that the covalent modifications on the parental H2A-H2B complex are diluted by half in the reassembled histone octamer by a newly incorporated H2A-H2B counterpart. On the other hand, newly incorporated (H3-H4)2 tetramers have to establish appropriate modifications de novo. Many in vitro experiments demonstrated that the two H2A-H2B dimers are first released with the support of the FACT (facilitates chromatin transcription) chaperon.20 This is followed by the release of the (H3-H4)2 tetramer, which is more tightly bound to DNA than H2A-H2B, by another chaperon anti-silencing factor 1 (ASF1). It has been shown that FACT is recruited to the replication fork through a direct or indirect interaction with MCM2-7.22 However, whether the same processes take place in vivo remains to be established.
Incorporation of newly synthesized histones into the daughter nucleosomes is dependent on multiple histone chaperons (Figure 1Bs). These newly synthesized histones are covalently modified before incorporation into chromatin. The best characterized examples include diacetylation of lysines 5 and 12 on histone H4 (H4K5,K12ac) by histone acetyltransferase 1 (HAT1)23 and H3K9me1.24 These modifications are removed around the time of deposition onto the daughter DNA duplex and their functions remain elusive. The newly synthesized (H3-H4) dimer binds to ASF1,25 is transferred to CAF-1, and then loaded onto the daughter DNA duplex by CAF-1.13,26 This loading is tightly coupled with the DNA replication fork through the interactions between PCNA and CAF-1 and between ASF1 and MCM2-7.23 On the other hand, newly synthesized histones H2A and H2B form a H2A-H2B dimer without any modifications and are loaded onto the daughter DNA duplex with the support of the chaperon nucleosome assembly protein 1 (NAP1).27
Parental histones transferred onto the daughter DNA duplex serve as templates to duplicate the same modifications by recruiting their respective modifying enzymes (Figure 1C). Several prominent histone modifying enzymes possess both a recognition and catalytic domain within the same protein molecule or complex. This dual capacity has the advantage of duplicating the same histone modification to nearby histones once a seed modification is provided. For example, the histone methyl transferase mixed lineage leukemia 1 (MLL1) induces the euchromatin marker H3K4me3 and binds to this same modification through the plant homeodomain (PHD) finger domain within MLL1.28 The heterochromatin marker H3K9me3 is mediated by suppressor of variegation 3-9 homolog 1 (Suv39H1) and Suv39H229 and recognized by HP130,31 which interacts with Suv39H1.32 The SETDB1–H3K9me3–HP1 connection mentioned earlier could also reinforce this interaction at the replication fork. Another heterochromatin marker, H3K27me3, is mediated by the enhancer of zeste homologue 2 (EZH2) subunit in polycomb repressive complex 2 (PRC2) and recognized by another subunit embryonic ectoderm development (EED).33 While these examples illustrate how a given histone modification can be inherited on daughter histones around the DNA replication fork, little is known about how these self-propagating histone modifications terminate excessive extensions to neighboring histones.
The study of the field of epigenetic maintenance of cell identity during replication is even more significant due to recent advances in the cell reprogramming field that challenge the cells identity. This link between DNA replication and cell differentiation/dedifferentiation has been a major question in stem cell biology. A classic example is the activation of oocyte-specific genes in somatic nuclei injected into Xenopus oocytes.34 Because the oocyte cytoplasm does not induce DNA replication in the injected nuclei, activation of oocyte-specific genes in the injected somatic nuclei was interpreted as evidence for the change of cell fate independently of DNA replication. More recently, it was shown that the efficiency of forming induced pluripotent stem (iPS) cells is dependent on the number of cell cycles they underwent.35 Other works indicate that a rapidly proliferating cell population is the primary source for iPS cells in a given culture dish.36,37 The speed of proliferation could be related to metabolism, senescence, and other cell physiology; however, the number of DNA replications in these experiments may also be an important determinant for the efficient change of the epigenetic state of parental cells by providing a vulnerable time-window for erasure of epigenetic memory. A more thorough understanding of the mechanism underlying the preservation of epigenetic memory during DNA replication would contribute to the effective maintenance and differentiation of stem cells as well as more efficient cell reprogramming.
N.K. was supported by the NIH (R01 GM098294, R21 CA187232, and R21 AR066158); Engdahl Family Foundation, Adjacent Possible Grant from the Stem Cell Institute, University of Minnesota and Grain-in-Aid of Research, Artistry and Scholarship, University of Minnesota (22802).
No potential conflicts of interests were disclosed.

©2015 Lowe, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.