Journal of
eISSN: 2373-437X


Research Article Volume 3 Issue 2
1Nucleo de Bacteriologia, Laboratorio Central de Saude Publica do Distrito Federal, Brazil
2Centro de Analises Proteomicas e Bioquimicas, Universidade Catolica de Brasilia, Brazil
3Faculdade de Ceilandia, Universidade de Brasilia, Brazil
Correspondence: Celio de Faria Junior, Nucleo de Bacteriologia, Laboratorio Central de Saude Publica do Distrito Federal, SGAN quadra 601, lotes O/P, Asa Norte, CEP: 70830-010, Brasilia, Brazil , Tel 55-61-3321-0774
Received: September 13, 2015 | Published: January 29, 2016
Citation: Faria-Junior C, Rodrigues LDO, Carvalho JOD, Franco OL, Pereira AL, et al. (2015) NDM-Producing Enterobacteriaceae Strains among Hospitals in BrasÃlia, Brazil. J Microbiol Exp 3(2): 00083. DOI: 10.15406/jmen.2016.03.00083
Carbapenem-resistant Enterobacteriaceae (CRE) strains have spread worldwide frequently driven by clonal spread. Additionally, plasmid-borne carbapenemase genes (blaKPC and blaNDM) have broadened the variability of species expressing resistance to carbapenems. This study aimed to characterize the susceptibility profile and bla genes in CRE strains recovered between 2012 and early 2014 in hospitals in Brasília, Brazil. Eighty eight CRE strains recovered from 19 medical settings were analyzed. Klebsiella pneumoniae positive for blaKPC accounted for the most of the CRE isolates (n=47; 53.4%). Seven blaNDM-positive strains (including K. pneumonia, n=4; Proteus mirabilis, n=1; Escherichia coli, n=1; and Providencia rettgeri, n=1) were recovered from patients in six hospitals. The first detected blaNDM-1-positive strain was P. rettigeri. Thereafter, blaNDM-1-positive K. pneumoniae strains showing indistinguishable Random Amplified Polymorphic DNA (RAPD) profiles were recovered in three hospitals. The susceptibility profile of blaNDM-1–positive K. pneumoniae strains was commonly restricted to amikacin, aztreonam and tigecycline. These dates highlighted the emergence of blaNDM-1–positive K. pneumonia strains marked by a single RAPD type among hospitals in Brasília, Brazil.
Keywords: ndm-producing strains, carbapenemase, klebsiella pneumonia, metallo-β-lactamase
The resistance to carbapenems has become a serious world public health issue since the early 2000’s.1 In that time, the world spread of Klebsiella pneumoniae carbapenemase (KPC)-producing strains was supported by the predominance of a well-adapted clone of K. pneumoniae (ST258) among hospitals around the world. Moreover, the blaKPC gene became easily mobilized by conjugative plasmids among Enterobacteria species.2 NDM-1 (New Delhi metallo-β-lactamase-1) is the most recently discovered molecular class B β-lactamase encoded on transferable, plasmid-borne genes (blaNDM).3 The hydrolysis mechanism of NDM relies on the interactions between β-lactam molecules and zinc ions in the enzyme’s active site. Therefore, NDM enzymes are inhibited by zinc-chelating agents such as EDTA.4 NDM can hydrolyze all β-lactam antibiotics (penicillins, cephalosporins and carbapenems), excepting monobactams.3 Additionally, most NDM-positive strains are broadly resistant to other antibiotic classes, and carry a wide diversity of resistance mechanisms against other antibiotics, such as aminoglycosides and fluoroquinolones, rendering these strains extremely resistant to the available treatments.1 NDM-1 was first described in K. pneumoniae and Escherichia coli strains isolated in Sweden in 2008 from an Indian patient who had been transferred from a hospital in New Delhi, India.5 Nowadays, NDM has also been detected in a broad variety of other Enterobacteriaceae species including K. oxytoca, Proteus mirabilis, Enterobacter cloacae, Citrobacter freundii and Providencia spp as well as in aerobic bacilli such as Pseudomonas spp. and Stenotrophomonas spp.3 This wide distribution of the blaNDM gene reflects its association with promiscuous plasmids.6 Regardless the purposes, whether medical or otherwise, international travels have played a significant role in the dissemination of NDM producers, given that, most of the first reports on NDM-positive strains were epidemiologically linked to travels to Indian and Pakistani regions.7 In Brazil, the first NDM-producing strain was isolated in 2013 in the South region state, Rio Grande do Sul.8 Beside blaKPC and blaNDM, other carbapenemase genes, such as blaVIM, blaIMP and blaOXA-48, have been reported in Enterobacteria world-wide, including in Brazil. However, these carbapenemase genes have not been associated with large spreads or epidemic events.9
The aim of this study was to define the profile of carbapenemase genes in CRE strains assessing whether NDM-producing strains have reached hospitals in Brasília, the federal capital of Brazil. Moreover, the study evaluated the role of bacterial clones in spreading of blaNDM among hospital.
From 2012 to 2014, a regional surveillance program was conducted by the Public Health Laboratory (LACEN-DF) in order to assess carbapenem resistance in Enterobacteriaceae isolates recovered from hospitals in Brasília. We have identified 88 carbapenem-resistant Enterobacteriaceae (CRE) strains recovered from patients attended in nineteen medical centers. Identification and antimicrobial susceptibility tests were accomplished using the MicroScan WalkAway™ system (Dade Behring, USA) and Vitek MS system (Matrix-assisted laser desorption ionization-time of flight mass spectrometry - MALDI-TOF MS system - BioMerieux) in accordance to the manufacturer’s instructions. In order to assess the clinical susceptibility of bacterial isolates, in vitro antibiogram test results were interpreted in accordance to the breakpoints established by the Clinical and Laboratory Standards Institute (CLSI) document published in January 2014. The production of carbapenemase was tested using the modified Hodge Test (MHT) employing ertapenem disk adsorbed with 10µg of the antibiotic as described by CLSI.10 Metallo-β-lactamase production was tested with carbapenem-containing disks (10µg meropenem or imipenem) adsorbed with 100mM EDTA.11 Control disks containing only carbapenem were used to evaluate the enlargement of inhibitory zones attributed to the EDTA effect. Specific primers were used in standard polymerase chain reactions (PCR) to detect the following carbapenemase genes: blaKPC (F, 5`TGTCACTGTATCGCCGTC and R, 5`CTCAGTGCTCTACAGAAAACC),12 blaNDM (F, 5`GGTTTGGCGATCTGGTTTTC and R, 5`GGCCTTGCTGTCCTTGATC), blaIMP (F1, CATTTCCATAGCGACAGCAC; F2, 5`AACACGGTTTGGTGGTTCTT and R, 5`GGACTTTGGCCAAGCTTCTA), blaVIM (F, 5`GATGGTGTTTGGTCGCATATC and R, 5`CTCGATGAGAGTCCTTCTAGAG) and blaOXA-48 (F, 5`GCGTGGTTAAGGATGAACAC and R, 5`ATCATCAAGTTCAACCCAACC). The primers used for amplification of blaNDM, blaIMP, blaVIM and blaOXA-48 were described in this study. PCR products were submitted to DNA sequencing (ABI 3130 Genetic Analyzer- Applied Biosystems®) in order to confirm the identity of the amplified genes. Clonal relatedness among isolates was examined by Random Amplified Polymorphic DNA (RAPD) performed with the primer OPA-2 (5’TGCCGAGCTG) (Operon Technologies, Alameda, CA, USA).13 The band patterns were analyzed by visual interpretation, applying the criteria established by Belkum et al.14 In addition, RAPD patterns were analyzed and dendograms were built employing PyElph software system (version 2.6.5).15
Among Enterotacteriaceae strains reported to the Public Health Laboratory (LACEN-DF) in Brazil, K. pneumonia was the most frequently CRE detected (n=61/88; 69.3%), followed by species of Enterobacter (n=15/88; 17.1%).
The emergence of CRE species has increased the demand on old or outdated antibiotics.16 In this scenario, the increasing interest on colistin (polymyxin) as an evaluable treatment has driven the emergence of species intrinsically resistant to colistin including Proteuss spp., Serratia spp., Morganella spp. and Providencia spp.17 In our study, intrinsically colistin-resistant strains accounted for 9.0% of the CRE isolates and they included S. marcescens (n=4/88; 4.5%), P. mirabilis (n=3/88; 3.4%) and P. rettgeri (n=1/88; 1.1%).
In relation to carbapenemase genes, blaKPC was the most frequently detected gene in the tested CRE strains (n=59/88; 67.0%), followed by blaNDM (n=7/88; 8.0%). Additionally, carbapenemase genes with minor epidemiological relevance were also tested (blaVIM, blaIMP and blaOXA-48), but they were not detected among the CRE isolates. Focusing on NDM genes, we firstly isolated a blaNDM-positive P. rettgeri strain from a necrotic ulcer affecting a 75-year-old male patient in May 2013. The patient had received treatment in two hospitals, both located in Brasília, and had not reported travelling abroad in the six previous years. The P. rettgeri strain showed in vitro resistance to all tested antimicrobial agents with the exception of gentamicin. The sequence analysis (Basic Local Alignment Search Tool) of the blaNDM amplicon showed an identity of 100% (435/435 base-pairs) with previous reported blaNDM-1 genes (GeneBank Number: KJ150691.1). Additionally, two distinct bands of plasmid DNA were found in the P. rettgeri strain (data not shown). PCR assays carried out with purified plasmid DNA showed that blaNDM-1 gene was located on the high-molecular-weight plasmid (molecular weight >50 Kb). Interesting, Carvalho-Assef et al.8 also recovered a NDM-producing P. rettgeri strain from a diabetic foot infection in early 2013, but differently the blaNDM-1 gene was chromosomally integrated.
Thereafter the first detection of blaNDM, six other blaNDM-positive strains (K. pneumonia, n=4; P. mirabilis, n=1; E. coli, n=1) were isolated in three hospitals. All isolates were resistant to β-lactams (with exception of aztreonam and cefotetan), quinolones, nitrofurantoin and trimethoprim-sulfamethoxazole; and showed variable susceptibility profiles against aminoglycosides, tetracycline and tigecycline (Table 1). Interesting, all NDM-producing strains showed negative results for the carbapenemase expression assay MHT. However, NDM-producing strains are positive in the EDTA test confirming the production of metallo-β-lactamases (Table 1). Negative or weakly positive results in MHT have been already reported for NDM-producing strains.18 However, these findings are worrisome once phenotypic detection of carbapenemase in MHT is recommended for the clinical microbiology laboratories as epidemiological screening assay for detection of CRE isolates.10
|
Patient |
Species |
Susceptible phenotypea |
Hospitals |
Isolation Date |
Assays for carbapenemase detection |
||
|
Phenotypic assay for carbapenemase (MHT) |
Phenotypic assay for metallo-β-lactamase (EDTA test) |
PCR for blaNDM |
|||||
|
1 |
Providencia rettgeri |
GEN, TET |
A |
03/06/2013 |
Negative |
Positive |
Positive |
|
2 |
Klebsiella pneumoniae |
AMI, AZT, TET, TGN |
B |
20/08/2013 |
Negative |
Positive |
Positive |
|
3 |
Klebsiella pneumoniae |
AMI, TGN |
B |
07/09/2013 |
Negative |
Positive |
Positive |
|
4 |
Proteus mirabilis |
AMI, CTE, GEN, TOB, |
B |
30/10/2013 |
Negative |
Positive |
Positive |
|
5 |
Klebsiella pneumoniae |
AMI, TGN |
C |
3/11/2013 |
Negative |
Positive |
Positive |
|
6 |
Klebsiella pneumoniae |
AMI, AZT, TET, TGN |
D |
11/11/2013 |
Negative |
Positive |
Positive |
|
6 |
Escherichia coli |
AMI, AZT, GEN, NIT, TGN |
E |
21/01/2014 |
Negative |
Positive |
Positive |
Table 1 NDM-producing strains isolated in Brasília hospitals
aAbbreviations: AMI, amikacin; GEN, gentamicin; TOB, tobramycin; TET, tetracycline; TGN, tigecycline; AZT, aztreonam; CTE, cefotetan
Four strains of NDM-producing K. pneumoniae were isolated from patients treated in three hospitals; therefore, it was tested if these strains were clonally unrelated as commonly reported for blaNDM-positive strains.19-21 However, all NDM-producing K. pneumoniae strains tested in this study were considered as genetically indistinguishable on RAPD analyses, showing the same amplified polymorphic DNA pattern (Figure 1B & 1C). Two of these strains were isolated from two patients (patients 2 and 3) assisted in the same hospital (hospital B), warning for the possibility of cross infections (Table 1 & Figure 1). The other two clonal strains of K. pneumoniae were isolated from two patients (patients 5 and 6) treated in two different hospitals (hospital C and D) (Table 1 & Figure 1). Moreover, because of a prolonged colonization period (2 months and 10 days) with blaNDM-positive strains (K. pneumoniae and E. coli), the patient 6 had the opportunity of translocating two NDM-producing enterobacterial species into two different hospitals (Table 1). Additionally, the isolation of different bacterial species positive for blaNDM from the patient 6 (Table 1) endorses the idea on the promiscuous nature of mobile genetic elements carrying blaNDM genes.3 These findings reinforce the role of patient transfer in spreading NDM-producing bacteria among hospitals,22 and endorses the need of rapid communication that alerts about the presence of infected or colonized patients with NDM-producing strains in Brazil hospitals.
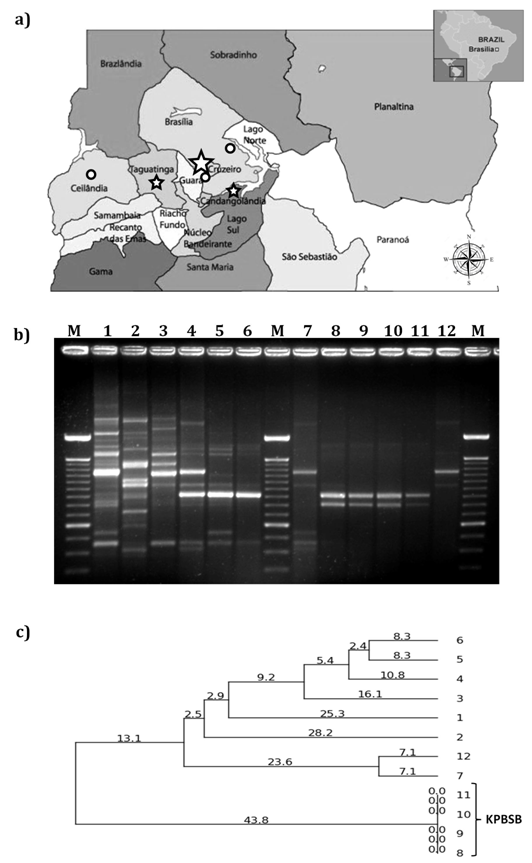
Figure 1 Distribution of cases of NDM-producing Enterobacteriaceae strains in Brasília and genetic relatedness of blaNDM-producing K. pneumoniae strains. A) Geographic distribution of the occurrence of NDM-producing strains. Symbols: Stars correspond to the cases associated with blaNDM-positive K. pneumoniae (small star - 1 case; large star - 2 cases). Circles indicate cases associated with other blaNDM-positive strains (Providencia rettgeri, P. mirabilis and Escherichia coli). B) RAPD profiles of the carbapenem-resitant K. pneumoniae strains. blaNDM-positive K. pneumoniae showed the same RAPD profile (samples 8-11) and were named KPBSB clone. Sample 1, K. pneumoniae IOC4955; sample 2, K. pneumoniae ATCC700603; samples 3 to 7 and 12 blaKPC-positive strains isolated in different hospitals in Brasília (enrolled to examine the discriminatory power of the RAPD assay); samples 8 and 9, blaNDM-positive strains isolated in hospital B; sample 10, blaNDM-positive strain isolated in hospital D; and, sample 11, blaNDM-positive strain isolated in hospital C. C) Dendrogram of the RAPD profiles as analyzed with PyElph software system (version 2.6.5). (see description of figure 1B for sample consultation).
As occurs in Brasília, Brazilian hospitals have frequently reported outbreaks involving CRE strains mainly associated with blaKPC-positive K. pneumoniae strains belonging to the clonal complex 258 (ST 11).23,24 The clone ST11 of K. pneumoniae has been characterized for causing large outbreaks,25 and has been also responsible for spreading blaNDM gene in Greece.26 Taken together, these data warn about the possibility of a worst-case scenario, in which, the epidemic clone ST11 would acquire the blaNDM gene and spread among Brazilian hospital.
Hospitals in Brazil have reported the isolation of several species of CRE positive for blaKPC, and, more recently, for blaNDM as well. Additionally, the initial spread of blaNDM-positive K. pneumoniae strains has been driven by a single clone. Our findings suggest that blaNDM-positive strains are been transported among hospitals by inpatient transfers and that they are spreading throughout patient cross infections. Finally, the present results call for an improved surveillance on inpatient transfers, for the molecular detection of CRE strains, and for enforcements in infection control measures.
All authors declare to have no conflict of interest.
Brasília Study Group on Bacterial Resistance: Alessandra Peres Pinheiro Domingues, Alessandra Reis Moreira, Brenda Paula Pires-Sousa, Eli Mendes Ferreira, Leonardo Borges Ferreira, Luana A A Martins, Melissa Jordão Sacramento, Michelle Capucci Martins. This work was supported by Fundação de Apoio à Pesquisa do Distrito Federal (Grant numbers: 193.000.019/2012, 2010/00188-1, 563981/2010-5), Conselho Nacional de Desenvolvimento Científico e Tecnológico (Grant number 301156/2011-5) and Coordenação de Aperfeiçoamento de Nível Superior (Grant number 001-2009).

©2016 Faria-Junior, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.