Journal of
eISSN: 2473-0831


Research Article Volume 2 Issue 2
Correspondence: Abdul Aziz MMEl Attar, Pharmaceutical Analytical Chemistry Department, Faculty of Pharmacy, Al-Azhar University, 11751 Nasr City, Cairo, Egypt
Received: October 25, 2015 | Published: March 29, 2016
Citation: Attia KA, Nassar MW, Attar AAM (2016) Different Aspects in Partial Least Squares and Artificial Neural Network Models Used for the Analysis of Cefoperazone Sodium in Presence of its Alkaline Degradation Product. J Anal Pharm Res 2(2): 00015. DOI: 10.15406/japlr.2016.02.00015
Several chemometric models were used for the determination of cefoperazone sodium in presence of its alkaline degradation product. The methods are either traditional (Partial Least Squares) or advanced (Artificial Neural Network). Partial Least Squares method was used with and without variable selection (Genetic Algorithm GA). Artificial Neural Network (ANN) was used with and without variable selection procedure (Genetic Algorithm GA) and data compression procedure (principal component analysis PCA). The chemometric methods used are PLS-1, GA-PLS-1, ANN, GA-ANN and PCA-ANN. The methods were used for the determination of cefoperazone sodium in bulk powder and pharmaceutical preparation. A 2-factor 5-level experimental design was built leading to 25 mixtures containing different ratios of cefoperazone sodium and its alkaline degradation product. Thirteen mixtures were used as a training set and the other twelve were used as a validation set.
Keywords:cefoperazone, PLS-1, GA-PLS-1, ANN, GA-ANN, PCA-ANN
Cefoperazone is sodium (7R)-7-((R)-2-(4-ethyl-2,3-dioxopiperazin-1 ylcarboxamido)-2-(4-hydroxyphenyl) acetamido)-3-((1-methyl-1H-tetrazol-5-yl) thiomethyl)-3-cephem-4-carboxylate. Its molecular weight is 667.6 and its molecular formula is C25H26N9NaO8S2. Cefoperazone is a third generation cephalosporin antibiotic, beta-lactam and inhibitor of cell wall synthesis. Cefoperazone (CPZ) is commonly used for infections caused by Gram-negative bacteria.1,2 Literature survey reveals that CPZ was determined in pure form, pharmaceuticals and biological fluids using spectrophotometric,3-12 voltammetric,13-16 chromatographic17-21 and fluorimetric22 methods (Figure 1).
The rationales of this work were to:
ANN is a kind of information processing chemometrical technique. It simulates some properties of human brain i.e. the way the input data are treated by the artificial (computer simulated) neuron is similar in action to a biological neuron exposed to incoming signals from neighboring neurons. In the computer the neurons are represented as weight vectors. Artificial Neural Network (ANN) applied in the field of regression or classification. In this manuscript ANN has been applied to establish a correlation between relationship between inputs and outputs. ANNs are composed of some units (input, hidden and output) and connection weights between the units. The neural networks where information flows from the input to the output layer are frequently termed ‘feed-forward’ ANNs (i.e. the type of ANN used in this manuscript is feed-forward network trained with the back propagation of errors learning algorithm). It is called feed-forward ANN as information passes one way through the network from the input layer, through the hidden layer and finally to the output layer. The outputs (predicted concentrations), are compared with targets (actual concentrations), and the difference between them is called error.23
The transfer functions
There are two transfer functions used in ANN; one between input and output of a node in the hidden layer and the other is applied in output layer. The use of these functions depends on relationship between the inputs and the outputs. Tansig-purelin transfer functions are commonly used for non-linear systems,24 while purelin-purelin functions are used for linear ones.25
Hidden neurons number (HNN)
It is related to the converging performance of the output error function during the learning process.
Number of neurons
Unfortunately, there are no fixed rules as to how many neurons should be included in the hidden layer. If there are too few nodes in the hidden layer the network may have difficulty generalizing to problems it has never encountered before. On the other hand, if there are too many nodes in the hidden layer, the network may take an unacceptably long time to learn anything of any value.
Lc, Lcd and Lci
The learning coefficient (Lc) controls the degree at which connection weights are modified during the learning process. The learning coefficient decrease (Lcd) and learning coefficient increase (Lci) control the variation of Lc value. It varies as a function of performance of the ANN.
Materials and reagents
Instruments
SHIMADZU dual beam UV-visible spectrophotometer (Kyoto/ Japan), model UV-1650 PC connected to IBM compatible and aHP1020 laser jet printer. The bundled software, UV-Probe personal spectroscopy software version 2.1 (SHIMADZU) was used. The spectral band was 2 nm and scanning speed is 2800 nm/min and 1 nm data interval.
Software
All chemometrics methods were implemented in Mat lab 8.2.0.701 (R2013b). PLS, GA-PLS, ANN, GA-ANN and PCA-ANN were carried out using PLS toolbox software version 2.1 in conjunction with Neural Network toolbox. The t-test and F-test were performed using Microsoft Excel.
Standard solutions
Spectral characteristics of CPZ and its degradate
The zero order (D0) absorption spectra were recorded against distilled water as a blank over a range of 200-400nm.
Experimental design for chemometric methods
A 5-level, 2-factor design was performed using 5 concentration levels for the drug and its alkaline degradate to be analyzed. The design spans the mixture space fairly well; where there are 5 mixtures for each compound at each concentration level, resulting in 25 mixtures.26 The central level of the design is 10µg/mL for both. Table 1 represents the concentration design matrix. The regions from 200 to 210nm and from 300 to 400nm were rejected. Thirteen mixtures of this design were used as a calibration set and the other 12 mixtures were used as a validation set to test the predictive ability of the developed multivariate models.
|
Mix. No |
Cefoperazone |
Degradate |
|
1a |
10 |
10 |
|
2 |
10 |
8 |
|
3 |
8 |
8 |
|
4 |
8 |
12 |
|
5 |
12 |
9 |
|
6 |
9 |
12 |
|
7 |
12 |
10 |
|
8 |
10 |
9 |
|
9 |
9 |
9 |
|
10 |
9 |
11 |
|
11 |
11 |
12 |
|
12 |
12 |
11 |
|
13 |
11 |
10 |
|
14 |
10 |
12 |
|
15 |
12 |
12 |
|
16 |
12 |
8 |
|
17 |
8 |
11 |
|
18 |
11 |
8 |
|
19 |
8 |
10 |
|
20 |
10 |
11 |
|
21 |
11 |
11 |
|
22 |
11 |
9 |
|
23 |
9 |
8 |
|
24 |
8 |
9 |
|
25 |
9 |
10 |
Table 1 The 5-level, 2-factor experimental design shown as concentrations of mixture components in μg/mL.
aShadowed rows represent the calibration set.
Analysis of Cefozon® by the proposed methods
Contents of 5 Cefozon® vials (1000mg/vial) were mixed well. An accurately weighed amount equivalent to 10mg of cefoperazone sodium was transferred into 100-mL volumetric flask. Cefoperazone sodium is dissolved in about 50mL distilled water, sonicated for 15min, diluted to the mark with distilled water mixed well and filtered; the first portion of the filtrate was rejected. The solution labeled to contain 100 μg/mL of cefoperazone sodium. Repeat the general procedure using aliquots covering the working concentration range. Determine the content of the vials from the corresponding regression equation. The spectra of these solutions were scanned from 200 to 400nm, stored in the computer and analyzed by the proposed methods.
Figure 2 shows the zero order UV absorption spectra of cefoperazone and its alkaline degradation product. The spectral overlapping of the drug and its degradation product prevents resolution of the mixture by the direct spectrophotometric measurements.
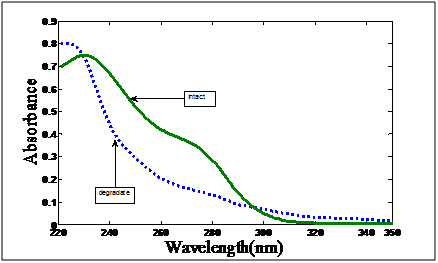
Figure 2 Zero order absorption spectrum of 24μg/mL cefoperazone and 24μg/mL its alkaline degradation product using distilled water as blank.
The aim of this study was to develop accurate and simple chemometric methods for determination of CPZ in presence of its degradate and to show the effect of data compression and variable selection on improving the predictive power of PLS and ANN models.
The first step in model building, involves constructing the calibration matrix for intact and its degradate. In this study the model was optimized with the aid of the 5-level 2-factor design26 resulting in 25 sample mixture. Table 1 shows the composition of the 25 sample mixtures. These 25 sample mixtures were divided to 13 training mixtures (for building the models) (odd numbers) and 12 validation mixtures (for measuring predictive power of the models) (even numbers).
The quality of multi component determination depends on the wavelength range and spectral mode used.27 The wavelengths used were in the range 221-350nm. Wavelengths less than 221 nm were rejected due to the noisy content. Wavelengths more than 350nm were not used because they were uninformative (no absorption is mentioned in these regions).
Variable selection: genetic algorithm (GA)
GA is used for optimization and for other applications such as wavelength selection in spectroscopy. Molecular spectroscopy has been greatly improved by the use of variety of multivariate statistical methods.28,29 Methods such as Partial Least Squares (PLS) or Principal Component Regression (PCR), allow taking into account the whole spectrum without performing variable selection.30 It has been recognized that an efficient variable selection can be beneficial to improve the predictive ability of the model and to reduce its complexity.31 Several techniques devoted to variable selection in PLS models applied to spectral data have been presented.32,33 It has already been shown that genetic Algorithms (GAs) can be successfully used as a variable selection technique.34,35 The architecture of a GA can be divided into five components: Initiation, Evaluation, Exploitation, Exploration and Mutation. An important issue of successful GA performance is the adjustment of GA parameters.36
Variable selection can be seen as an optimization problem. Among the different variable selection algorithms available, Genetic Algorithms (GA) are the most commonly employed. The main idea behind the use of GA in numerical optimization is the mathematical translation of the biological concept of the ‘survival of the fittest’. The fitness values were used as response variables. Mutation rate was 0.005 in all cases as when it increased above this value, no convergence occurred between average fitness and best fitness values and model stop. The adjustment of the GA parameters is shown in Table 2.
|
Parameter |
Value |
|
Population size |
20 |
|
Maximum generations |
38 |
|
Mutation rate |
0.005 |
|
The number of variables in a window(window width) |
2 |
|
Per cent of population the same at convergence |
100 |
|
%Wavelengths used at initiation |
50 |
|
Crossover type |
double |
|
Maximum number of latent variables |
2 |
|
Cross validation |
Random |
|
Number of subsets to divide Data into for cross validation |
4 |
|
Number of iterations for cross validation at each generation |
2 |
Table 2 Parameters of the genetic algorithms
Each solution (chromosome) is evaluated using the PRESS value reached in the calibration. The genetic algorithm searches for the minimum PRESS in the space of all the possible chromosomes without establishing, a priori, the latent structure of the calibration.
Where and are predicted and true concentrations in µg/mL, respectively. The GA was run for 129 variables (in the range 221- 350) for cefoperazone. The selected variables (86) were used for running of PLS model and ANN. GA reduced absorbance matrix to about 34-36 % of the original matrix.
Partial least squares (PLS-1)
The purpose of PLS method is to build a calibration model between the concentration of the components under study (CPZ and DCPZ) and the latent variables of the data matrix.37,38 Two different aspects can be used in Partial Least Squares called PLS-1 and PLS-2. PLS-2 uses the whole information about the concentration of all components to form latent variables (LVs), while PLS-1 uses only the information about the concentration of one component to create the LVs used by the model.38
Including extra LVs in the model increases the possibility of the known problem of over fitting. On the other hand, if the number of LVs was too small meaningful data that could be necessary for the calibration might be discarded. Therefore, optimization of number of the LVs is a critical issue in PLS method. Leave one out (LOO) cross validation and the bootstrap39 can be applied to predict the optimum number of PLS components. PLS-1 calibration on 12 calibration spectra was performed and, using this calibration, the concentration of the sample left out during the calibration process was predicted. This process was repeated 13 times until each training sample had been left out once.40 The predicted concentrations of the components in each sample were compared with the actual concentrations in this calibration samples and the root mean squares error of cross-validation (RMSECV) was calculated as follows:
Where I is the number of objects in the calibration set, Yt is the known concentration for sample, and Ypredt is the prediction concentration of sample.
The RMSECV was used as a diagnostic test for examining the error in the predicted concentrations. It indicates both of the precision and accuracy of predictions.
Appropriate selection of the number of factors to be used to construct the model is a key to achieve correct quantitation in PLS-1 calibration. The most usual procedure for this purpose involves choosing the number of factors that result in the minimum RMSECV. However, this criterion is subjected to some constraints since, occasionally; the RMSECV does not reach a sharp minimum, but decreases gradually above a given number of factors. For this reason, the method developed by Haal et al.28 was used for selecting the optimum number of factors, which involves selecting that model including the smallest number of factors that results in an insignificant difference between the corresponding RMSECV and the minimum RMSECV (Figure 3a & 3b).

Figure 3a The optimum number of LV for cefoperazone sodium and it’s degradate concentration prediction from raw data.

Figure 3b The optimum number of LV for cefoperazone sodium and it’s degradate concentration prediction from GA model.
ANN
An ANN is a set of interconnected neurons (also termed nodes, cells, units or process elements) distributed in a specific arrangement, usually termed architecture. In general, neurons are organized in layers. The most common neural nets, the feed-forward nets, are fully connected, i.e. each node is connected to all the nodes in the next layer. The information we want to enter in the ANN (e.g. the spectra) is given to the ‘input layer’, which is the set of neurons that receive directly the information from the external world of the net. In inverse calibration, the inputs could be the absorbances at various wavelengths and the output could be the concentrations of one or more analyte. The ANN consists of three layers; two layers with connections to the outside world (an input layer where data are presented to the network and an output layer which holds the network response to given inputs ) and one hidden layer (optimized afterwards). Reduce the number of input variables without losing relevant information leads to time saving. A powerful tool to carry out such a reduction in the dimensionality of the original data was principal components analysis (PCA). We can consider that the scores of (PCs) alone represent the calibration standards. So, instead of using the full original absorbance variables to train the ANN, only a reduced number of PC scores can be used. Also, genetic algorithm was used to select the best wavelengths to represent each compound. This means that, the absorbance matrix was reduced either by Genetic Algorithm (variable selection procedure) to about 34% of the original matrix or Principal Component Analysis (PCA) (variable compression procedure) to three principal components. Thus, three ANNs (ANN, GA-ANN and PC-ANN) were applied in our work. The output layer is the concentration matrix of one component. The hidden layer consists of just single layer which has been considered sufficient to solve similar or more complex problems. Moreover, more hidden layers may cause over fitting.25
Optimization of ANN parameters is of a great importance for a proper modeling. These parameters are HNN, Lc, Lci. Placket- Burman design was used for Optimization as shown in Table 3. The choice of the proper transfer function depends on the nature of data.
|
Method |
ANN |
GA-ANN |
PCA-ANN |
|
Drug |
Cefoperazone |
Cefoperazone |
Cefoperazone |
|
Hidden Neurons Number |
10 |
10 |
10 |
|
Transfer Functions |
Purelin-Purelin |
Purelin-Purelin |
Purelin-Purelin |
|
Learning Coefficient |
0.01 |
0.01 |
0.01 |
|
Learning Coefficient Decrease |
0.1 |
0.1 |
0.1 |
|
Learning Coefficient Increase |
100 |
100 |
100 |
Table 3 Optimized parameters of ANNs
After optimization of parameters and architectures of the ANNs the training step is preceded. In other words, learning is the process by which an ANN modifies its weights and bias terms in response to the input information (spectra and concentration values). The error correction learning is known as back-propagation. This learning mode compares the outputs of the ANN with the true concentration values. The error derived from such a comparison will control the ANN training. The errors can be used to adjust the top weights directly by means of a predefined algorithm. Error-correction learning algorithms attempt to minimize error on each iteration. TRAINLM32 was thus preferred as it is time saving.
Since relationship between absorbance and concentration is linear, purelin-purelin (as a transfer function between input and hidden layer; and between hidden layer and outer layer) was found to give best results in our work.
The network is trained using the training set: in the calibration example the ANN would calculate concentrations for each member of the training set, and any discrepancies between the network’s output and the known concentrations would be used to adjust internal parameters in the network. These prediction and adjustment steps are repeated until the required degree of accuracy, evaluated with a test set, is achieved. Since the training and test sets are bound to differ to some extent, it is important not to over-fit the training set, otherwise the network may perform less well with the test set, and subsequently with ‘unknown’ samples.
ANNs are useful when the mathematical model is unknown or uncertain because they do not assume any mathematical relationship between the input and output variables. The proposed chemometric methods were run on the calibration data using optimal parameters. The concentrations of the drug and it’s degradate in the calibration set (13 mixtures) were calculated. In order to validate the proposed methods, the validation set (12 mixtures) was analyzed with the proposed methods (Table 4).
|
Concentration |
(μg/mL) |
PLS-1 |
GA-PLS |
ANN |
GA-ANN |
PCA-ANN |
|
Cefoperazone |
Degradate |
Recovery %a |
||||
|
10 |
8 |
99.4 |
101.9 |
101.1 |
101 |
102 |
|
8 |
12 |
101.13 |
101.88 |
99.25 |
98.88 |
98 |
|
9 |
12 |
101.36 |
101.33 |
98.89 |
99.89 |
97.78 |
|
10 |
9 |
101.16 |
99.1 |
98.71 |
99.5 |
98.7 |
|
9 |
11 |
100.51 |
98.78 |
98.39 |
99.89 |
98.33 |
|
12 |
11 |
98.51 |
98.92 |
98.83 |
99.92 |
98.83 |
|
10 |
12 |
101.95 |
99.9 |
98.61 |
99 |
98.6 |
|
12 |
8 |
98.81 |
99.17 |
98.33 |
99.95 |
98.33 |
|
11 |
8 |
99.82 |
101.11 |
101.13 |
100.19 |
101.27 |
|
10 |
11 |
98.05 |
101.6 |
101.06 |
100.6 |
101.6 |
|
11 |
9 |
98.11 |
99.16 |
99.91 |
99.91 |
97.18 |
|
8 |
9 |
98.02 |
99.38 |
99.38 |
99.75 |
98.75 |
|
Mean |
99.74 |
100.19 |
99.47 |
99.87 |
99.11 |
|
|
RSD% |
1.45 |
1.261 |
1.08 |
0.59 |
1.604 |
|
|
RMSEPb |
0.141 |
0.12 |
0.1195 |
0.0544 |
0.1769 |
Table 4 Determination of Cefoperazone sodium and its degradate in validation set by the proposed chemometric methods
aAverage of three determinations.
bRoot mean square error of prediction.
The proposed PLS-1, GA-PLS, ANN, GA-ANN and PCA-ANN methods were successfully used for the determination of CPZ in Cefozon vial, Table 5.
|
|
PLS-1 |
GA-PLS |
ANN |
GA-ANN |
PCA-ANN |
Reported Method |
|
Mean |
100.8 |
100.14 |
100.38 |
100.07 |
100.54 |
100.22 |
|
N |
5 |
5 |
5 |
5 |
5 |
5 |
|
SD |
1.131 |
0.821 |
0.675 |
0.791 |
1.241 |
1.679 |
|
Variance |
1.278 |
0.676 |
0.455 |
0.625 |
1.541 |
2.821 |
|
Student's t Test |
0.641 |
0.093 |
0.191 |
0.186 |
0.339 |
|
|
-2.306 |
-2.306 |
-2.306 |
-2.306 |
-2.306 |
||
|
F Value |
2.206 |
4.172 |
6.195 |
4.519 |
1.831 |
|
|
-6.388 |
-6.388 |
-6.388 |
-6.388 |
-6.388 |
|
Table 5 Statistical comparison for the results obtained the proposed methods and reported method16 the analysis of cefoperazone sodium in cefozon® vial
The results obtained for the analysis of CPZ in Cefozon vial by the suggested methods were statistically compared with those obtained by applying the reported second derivative method12 and no significant difference between the results was obtained as shown in (Table 5). Using one-way ANOVA test, the obtained results by applying these methods showed no significant differences among all of them as shown in Table 6.
|
Drug |
Source |
DF |
Sum of Squares |
Mean of Squares |
Value F |
|
Cefoperazone sodium |
Between exp |
5 |
1.83 |
0.367 |
0.302 (2.621) |
|
Within exp |
24 |
29.2 |
1.216 |
Table 6 One-way ANOVA test for the different proposed methods used for the determination of cefoperazone sodiumin Cefozon® vials
The values between parentheses are the theoretical F values.
The population means are not significantly different.
GA reduced the optimal number of latent variables of PLS-1 model for CPZ from three into two factors. Also, recoveries and RMSEP (Root Mean Square Error of Prediction) were decreased indicating a better resolution power of GA-PLS model than PLS-1 model (Table 4).
The comparison shows that GA-ANN is more suitable for the determination of cefoperazone because GA allows the use of less number of neurons (so shorter training time) for cefoperazone than those used in the networks utilized raw. While PCA-ANN did not show any improvement than ANN, even the results were worse (Table 4). These results indicate that variable selection models (GA) are more suitable than data compression procedure (PCA), when preceding ANN, for the analysis of this binary mixture. This result may be attributed to the fact that GA introduces the most relevant wavelengths to the drug concentration.
ANN is better than PLS-1 (Table 4), which may be due to the fact that ANN is a type of artificial intelligence and that in ANN there is no chance for over fitting that may occur in PLS calibrations.
In conclusion, the described chemometric methods gave accurate and precise results for determination of cefoperazone in presence of its alkaline degradation product without prior separation and can be applied for routine analysis.
None.
Authors declare that there is no conflict of interest.

©2016 Attia, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.