Journal of
eISSN: 2473-0831


Research Article Volume 9 Issue 1
1Department of Genetics, Sanjay Gandhi Post Graduate Institute of Medical Sciences, India
2Department of Neurology, Sanjay Gandhi Post Graduate Institute of Medical Sciences, India
Correspondence: Ashok Kumar, Department of Genetics, Sanjay Gandhi Post Graduate Institute of Medical Sciences (SGPGIMS), Lucknow-India-226014
Received: January 25, 2020 | Published: February 26, 2020
Citation: Kumar A, Agarwal S, Pradhan S. CpG methylation and various parameters interaction in myotonic dystrophy type 1. J Anal Pharm Res.2020;9(1):16?25. DOI: 10.15406/japlr.2020.09.00347
Myotonic dystrophy type 1 is a chronic, slowly progressing, inherited multisystemic autosomal-dominant disease, caused by expansion of CTG repeats in DMPK gene. The purpose of the present study was to analyze molecular expansion profiling of CTG repeat, status of CpG methylation at DMPK gene locus, and to established relationship between CpG methylation and CTG repeat expansion size along with other clinical and biochemical parameters. Clinically suspected 21 DM1 subjects, 56 family members and 50 normal individuals were included in this study. Molecular diagnosis of CTG repeat expansion was performed by Myotonic Dystrophy Short PCR (MDSP) and Triplet primed-PCR (TP-PCR) and followed fragment analysis on ABI-310 Genetic Analyser. The CpG methylation was done by bisulphite conversion kit (Cells to CpGTM Bisulphite conversion kit, 4445555) and 7500 Fast RT-PCR. SPSS version 16 and Pearson correlation coefficient were used for statistical analysis. All clinically suspected 21 subjects had CTG repeat expansion. Among 56 family members, 16 were permutated, and 40 were normal for CTG repeat. Our previous findings (Kumar et al, 2016 and 2018) highlighted that pattern of CTG repeats differs according to ethnicity. Among positive DM1 samples (n=21), 13 samples were methylated. CpG methylation was significantly correlated only with CTG repeat expansion. This methylation may affect the disease environment and expression of neighbor gene which is responsible for disease pathogenesis.
Keywords: Myotonic dystrophy type 1 (DM1), CTG repeat expansion, TP-PCR, RT-PCR, CpG methylation, bisulphate, myotonin protein kinase, muscular dystrophy, distal muscles
DM1, myotonic dystrophy type 1; UTR, untranslated region; FA, Friedreich ataxia; SCA, spinocerebellar ataxia; SCK, serum creatinine kinase; NCV, nerve conduction velocity; SSS, severity of sum score
Myotonic dystrophy type 1 (DM1) is the most common type of adult muscular dystrophy and inherited as autosomal dominant manner. It is characterized by myotonia, wasting of distal muscles, cataracts, cardiac arrhythmias, hypogonadism, frontal balding, and mental impairment etc.1 It is caused by an abnormal expansion of the CTG sequence located at the 3′ untranslated region (UTR) of the myotonin protein kinase gene (DMPK) at chromosome 19q13.3.2–4 It varies in normal population from 5 to 35 and beyond 50 repeat is responsible for DM1 severity.5–8 DMPK gene is ~14 kb and encodes 2.3 kb of mRNA with 15 exons and have 624 amino acids containing serine–threonine kinase protein.9,10
There are two precise mechanisms for disease pathogens of DM1. First, the expanded CUG repeats containing RNA expression alters the activity of RNA splicing factors and the consequence is that unregulated alternative splicing of many different genes.11 The other mechanism illustrate that the expanded CTG repeats interfere with local chromatin configuration and affects expression of both DMPK and neighboring genes DMAHP (dystrophia myotonica associated home domain protein)/SIX5, dystrophia myotonica containing WD repeat motif (DMWD), and muscle blind protein (MBNL1).12 Expanded CTG repeats interferes with the transcription of the adjacent DMAHP gene and thus play a role in disease manifestation.13 The decreased Six5 transcription is important in DM1’s pathophysiology as cataract formation.14 Additionally, anti-sense strand expression of DMPK also modified by expansion.15 Expanded DM1 repeats altered chromatin packaging at DM1 locus as compared to wild-type allele. This DM1 chromatin packaging is mediated by CpG methylation sensitive binding of the chromatin insulator CTCF proximal to the CTG repeat. Other diseases like fragile X syndrome (FRAXA), Friedreich ataxia (FRDA) etc. have differential CpG methylation pattern and chromatin packaging which manifest gene expression.16,17
Diagnosis of DM1 in clinical setup is difficult, because it contains similar clinical symptoms with spinocerebellar ataxia (SCA) and Friedreich ataxia (FA) and ther triplet repeat disorders. On clinical scale, myotonic dystrophy is diagnosed by the high level of muscle Serum creatinine kinase (SCK), characteristics electromyography (EMG) peaks, nerve conduction velocity (NCV), muscle biopsy, and several other variables. The variability in clinical features of DM1, require molecular testing for accurate diagnosis. Molecular methods used for detection of normal and expanded alleles of disease, are PCR-RFLP (Restriction fragment length polymorphism),18 Nested and fluorescent PCR,19,20 multiplex PCR and Southern blotting,21–23 and Triplet Primed-PCR (TP-PCR),24 and a choice methodology has been incorporated. In India, still the diagnosis is totally confined to clinical characterization with no prenatal and counseling set up.
Like various genetic markers, CpG methylation may also play role in the stability of repetitive sequence,25,26 and especially in behaviour of the CTG repeat,27,28 and in the pathogenesis of disease.27,29,30 Therefore, it is necessary to find out the molecular expansion profiling of CTG repeat (by normal and TP-PCR analysis), & status of methylation on CpG sites in the myotonic dystrophy locus, and to point out co-relation of CpG methylation with CTG repeat expansion size and other clinical and biochemical parameters.
Clinical evaluation
Institutional Ethics Committee of SGPGIMS approved this study. Patients were diagnosed from March 2016 to Dec 2018. Total 21 DM1 patients and 56 related individuals (peripheral blood) from 21 families were referred to SGPGIMS OPD & IPD and also 50 normal individual were diagnosed for CTG repeat diagnosis. No cell lines were used for the study. Patients having disorder of muscle wasting, jaw and temporal wasting, impairment in gripping capacity, arrhythmia, facial weakness and hypersomnia included in this present study while patients having any other neurological disorder or any other severe or familiar disease excluded from the present study. Clinically, DM1 patients evaluated by detailed disease history, age of onset, physical examination, biochemical testing and electrophysiological testing like NCV (nerve conduction velocity), and EMG (electromyography) and by several other variables. There is certain inclusion and exclusion criteria of the study are as follows:
Inclusion criteria for DM1- Clinical output of DM1, muscle wasting with arrthymia, muscle wasting with facial weakness, and impairment in gripping.
Exclusion criteria- Presence of any other neurological disorder, and presence of strong family history of congenital and severe disease except DM1
Grading of clinical severity
For evaluation a scoring system was adopted to assess the severity of the salient clinical features and this scale has provided 0 (minimum)–70 (maximum) points.8,31 The value 70 highlights severe illness (Table 1). In Indian scenario, clinical score ranging from 8-39 suggesting a mild to moderate severity of disease. The degree of muscle impairment was measured by a five point Muscular Disability Rating Scale (MDRS) measure.32
Molecular genetic analysis
Extraction of DNA samples
DNA isolation was processed by standard phenol chloroform method. The absorbance ratio at 260 and 280nm of DNA was around 1.7-1.9. The quality and purity was assessed by 0.8% agarose gel electrophoresis in 1X TBE buffer.
PCR and CTG repeat expansion analysis
Myotonic Dystrophy Short PCR (MDSP) was used to determine the sizes of normal and/or permutated triplet repeats. PCR was performed in a reaction volume of 25µl using 50ng genomic DNA with 5pmols of each primers 101-F (5’-FAM-CTT CCC AGG CCT GCA GTT TGC CCA TC-3’) and 102-R (5’-GAA CGG GGC TCG AAG GGT CCT TGT AGC-3’).3 The cycling profiles were: 5min at 95℃, 34cycles of 10sec at 95℃, 30sec at 62℃, and 30sec at 72℃. A final extension at 72℃ for 10min completed the reaction. Repeat size was calculated by subtracting the number of base pairs of the flanking region from the total length of the PCR products and dividing the result by three. ABI PRISM 310 Genetic Analyzer with the Gene Mapper ID 3.1 software (Applied Biosystems, Foster City, CA, USA) was used for fragment analysis.
For expanded CTG repeat allele expansion, Triplet Repeat PCR (TP-PCR) was carried out in a reaction volume of 25µl using 50ng genomic DNA with 10 pmol of primer P1-F (5’-FAM-AGA AAG AAA TGG TTC TGT GAT CCC-3’), 8pmol of primer P3-R (5’- TAC GCA TCC CAG TTT GAG ACG-3’), and 2 pmol of primer P4CTG (5’ TAC GCA TCC GAG TTT GAG ACG TGC TGC TGC TGC TGC T-3’).24 The reaction conditions were as: 10min at 96℃, 30 cycles of 1 min at 94℃, 1 min at 60℃, 2 min at 72℃, followed by final extension at 72℃ for 10 min. Again, the fragment analysis was done by ABI PRISM 310 Genetic Analyzer.
Assessment of repeat expansion
CTG repeat expansion carried out in all individuals (n=77) belonging from 21 DM1 family and also in 50 control subjects. Complete family screening was performed in twelve DM1 families (family 1, 3, 4, 8-11, 13-15, 17, 18).
CpG methylation at myotonic dystrophy locus
Bisulphite conversion
Bisulphite conversion kit (Cells to CpGTMBisulphite conversion kit, 4445555) was used for conversion of isolated DNA sample into bisulphite converted DNA as per manufacturer protocol. The PCR reactions were subjected to 2cycles of 65°C for 30min. and 95°C for 1.5min., and finally 65°C for 30min.
Real Time-PCR (RT-PCR)
The 7500 Fast RT-PCR was used for CpG methylation of myotonic dystrophy locus by using following set of
primers (F-5’AGAGGGAGTAGTAGTYGTTAGTTTAGTTT3’;R-5’ACTCCRATTTCCCTATAACCCAAA3’ and probes: Meth-[6 FAM]TCCCACCCTAAACGCCGACCTCTCCC[QSY] and Non- meth[HEX]TCCTCCCACCCTAAACACCAACCTCTCCC[QSY]. The PCR reactions were subjected to 40cycles of 95°C for 10sec., 52°C for 30 sec., and 72°C for 1min. followed by a 30sec. extension at 72°C.
Appropriate statistical analysis software (Student t test and Pearson/Spearman correlation) and SPSS (version 16) were used for analysis. A p value of <0.05 was statistically significant.
During the period of about three year (March 2016 to Dec 2018), blood samples (n=77) from 21 DM1 families had been collected at SGPGIMS. Among them 21 had pathogenic DM1, corresponds to 27.27% of total individual. Mean age at onset of symptoms, mean age of presentation (sampling age), mean duration of illness were 23.86±9.86 (range 1-38 year), 27.76±9.74 (range 8-40 year), 3.90±1.55 (range 2-7 year) respectively. Similarly, the no. of functionally independent, minimal help, dependent for daily activities, and wheel chair bounds patients were 5, 12, 3, 1 respectively (Table 1).
Patient/Sex |
Age at onset (y) |
Age at presentation (y) |
Dur(y) |
FH |
MT |
WS |
WK |
DM |
CD |
D |
ADL |
Asp |
T |
CK |
NC |
EMG |
Score |
MDRS |
7 |
6 |
15 |
3 |
6 |
6 |
6 |
6 |
6 |
6 |
1 |
2 |
70 |
||||||
1/F |
15 |
17 |
2 |
n |
3 |
4 |
8 |
- |
- |
- |
2 |
- |
- |
1 |
1 |
2 |
21 |
2 |
2/M |
11 |
14 |
3 |
p |
3 |
5 |
12 |
- |
- |
2 |
- |
- |
- |
1 |
1 |
1 |
25 |
3 |
3/M |
36 |
40 |
2 |
p |
4 |
4 |
6 |
- |
- |
- |
- |
2 |
4 |
- |
1 |
1 |
22 |
4 |
4/M |
18 |
20 |
2 |
m |
4 |
6 |
7 |
- |
- |
- |
2 |
- |
- |
- |
1 |
1 |
21 |
3 |
5/M |
20 |
22 |
2 |
n |
4 |
4 |
6 |
1 |
- |
2 |
- |
- |
- |
- |
1 |
- |
18 |
2 |
6/M |
25 |
30 |
5 |
p |
4 |
4 |
8 |
- |
- |
- |
- |
- |
2 |
1 |
1 |
1 |
21 |
2 |
7/M |
29 |
32 |
3 |
m |
4 |
5 |
7 |
- |
- |
- |
2 |
4 |
- |
1 |
- |
2 |
25 |
4 |
8/M |
36 |
41 |
5 |
m |
3 |
5 |
1 |
1 |
- |
- |
2 |
- |
- |
1 |
1 |
1 |
15 |
1 |
9/M |
23 |
26 |
3 |
m |
2 |
2 |
4 |
- |
- |
- |
2 |
- |
6 |
- |
1 |
2 |
19 |
2 |
10/M |
38 |
40 |
2 |
m |
3 |
2 |
4 |
- |
- |
- |
4 |
- |
- |
- |
- |
1 |
14 |
3 |
11/M |
25 |
28 |
3 |
m |
4 |
5 |
6 |
- |
- |
2 |
2 |
2 |
- |
1 |
1 |
- |
23 |
2 |
12/M |
21 |
25 |
4 |
p |
4 |
4 |
7 |
2 |
- |
- |
2 |
4 |
- |
1 |
1 |
2 |
27 |
3 |
13/M |
28 |
31 |
3 |
p |
7 |
6 |
8 |
- |
- |
- |
- |
- |
4 |
- |
- |
1 |
26 |
3 |
14/M |
13 |
18 |
5 |
m |
4 |
5 |
6 |
2 |
4 |
- |
4 |
2 |
- |
- |
1 |
1 |
29 |
5 |
15/M |
10 |
15 |
5 |
n |
4 |
4 |
8 |
- |
2 |
- |
2 |
2 |
6 |
1 |
1 |
2 |
32 |
2 |
16/M |
26 |
32 |
6 |
m |
2 |
5 |
2 |
1 |
4 |
4 |
6 |
- |
- |
1 |
1 |
1 |
27 |
1 |
17/M |
31 |
38 |
7 |
p |
3 |
4 |
4 |
1 |
2 |
2 |
2 |
- |
- |
- |
1 |
2 |
21 |
3 |
18/M |
30 |
34 |
4 |
m |
3 |
5 |
3 |
- |
- |
- |
2 |
- |
4 |
- |
1 |
1 |
19 |
2 |
19/F |
36 |
40 |
4 |
n |
5 |
5 |
7 |
- |
- |
2 |
4 |
2 |
- |
1 |
1 |
2 |
29 |
3 |
20/M |
29 |
32 |
3 |
p |
4 |
4 |
1 |
- |
2 |
2 |
2 |
2 |
- |
3 |
1 |
1 |
22 |
1 |
21/F |
1 |
8 |
7 |
n |
4 |
5 |
6 |
- |
- |
- |
2 |
- |
- |
1 |
1 |
1 |
20 |
2 |
Table 1 Clinical features and severity scale and scores in 21 DM1 patients
F, Female; M, Male; Dur, Duration of the disease; FH, Family history; n, Unknown; p, paternal; m, maternal; MT, Myotonia; WS, Wasting; WK, Weakness; DM, Diabetes Mellitus; CD, Cardiac Involvement; D, Dysphasia; ADL, Activities of Daily living; Asp., Aspiration; T, Testicular Atrophy; CK, Creatinine Kinase; NC, Nerve Conduction; EMG; Electromyography; MDRS, Muscular Disability Rating Scale. The maximum score (70) of all parameters were
Investigations highlighted normal CK (below 192 U/L), between 192-500U/L, and over 1000U/L in 12, 11, 1 patients respectively. Nerve conduction was abnormal in 18 (85.71%) and myopathic EMG limited to distal muscles in 12 (57.1%) and to proximal muscle in addition to distal muscle in 7 patients (33.3%). The clinical score or severity of sum score (SSS) had varied from 14-32. The 6 patients having a score between 14-20, 14 between 21-30, and 1 between 31-32 (Table 1).
Total 21 patients and 56 related individuals were recruited in the present study for molecular diagnosis of the disease by MDSP and TP-PCR methodology3,8,17 (Figure 1 & 2).
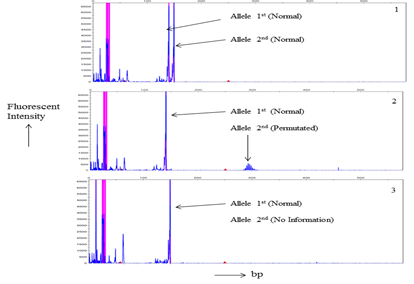
Figure 1 Myotonic Dystrophy Short-PCR (MDSP) product Gene scans analysis of a. Electrophorogram of a normal individual of a DM1 family member. b. an asymptomatic DM1’s individual electrophorogram. c. Picture of a DM1 patient. Horizontal scale indicates size in base pairs while fluorescent intensity of the normal and/or normal/permutated alleles represented by vertical scale.
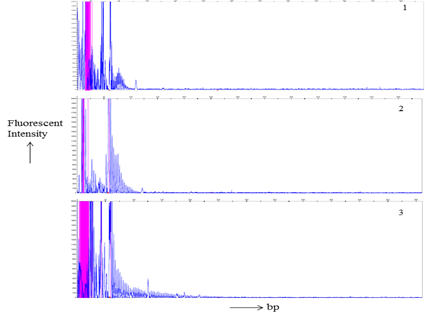
Figure 2 TP-PCR products Gene scan analysis of a. electrophorogram of a normal individual of a DM1 family member. b. an asymptomatic DM1’s individual electrophorogram. c. Picture of a DM1 patient. Horizontal and vertical scales indicate size in base pairs and fluorescent intensity of the expanded alleles respectively.
MDSP amplify normal and permutated allele only (Figure 1). TP-PCR provide specific amplification pattern for normal, permutated and expanded CTG repeat allele (Figure 2). Out of 21 patients; nine, seven, and five had acquired maternal, paternal and unknown mode of transmission of disease respectively (Table 1). Among 56 family members, sixteen were permutated (28.57%), and remaining (n=40) were normal (71.43%) for CTG repeat (Table 2). Total twelve families (family 1, 3, 4, 8-11, 13-15, 17, 18) undergo complete family screening.
DM1 Family(S.N.) |
Samples |
Normal PCR CTG repeats for allele 1/allele 2 |
TP-PCR Results |
Conclusion |
Family 1 |
Patient |
5/nd |
Expansion |
Full mutation |
Patient’s Mother |
11-May |
Normal |
Normal repeat |
|
Patient’s Brother |
16-May |
Normal |
Normal repeat |
|
Patient’s Sister |
16-May |
Normal |
Normal repeat |
|
Family 2 |
Patient |
3/nd |
Expansion |
Full mutation |
Patient’s Father |
16/pm |
Confirmed pre mutation |
Pre mutated allele |
|
Family 3 |
Patient |
16/nd |
Expansion |
Full mutation |
Patient’s Father |
16/pm |
Confirmed pre mutation Normal |
Pre mutated allele |
|
Patient’s Mother |
15-Nov |
Normal |
Normal repeat |
|
Patient’s Brother |
11-Mar |
Normal repeat |
||
Family 4 |
Patient |
11/nd |
Expansion |
Full mutation |
Patient’s Father |
16-Mar |
Normal |
Normal repeat |
|
Patient’s Mother |
11/pm |
Confirmed permutation |
Pre mutated allele |
|
Patient’s Sister |
11-Mar |
Normal |
Normal repeat |
|
Patient’s Brother |
11-Aug |
Normal |
Normal repeat |
|
Family 5 |
Patient |
16/nd |
Expansion |
Full mutation |
Patient’s Father |
16-May |
Normal |
Normal repeat |
|
Patient’s Sister |
16-Mar |
Normal |
Normal repeat |
|
Family 6 |
Patient |
16/nd |
Expansion |
Full mutation |
Patient’s Father |
16/pm |
Confirmed permutation |
Pre mutated allele |
|
Patient’s Mother |
15-May |
Normal |
Normal repeat |
|
Family 7 |
Patient |
18/nd |
Expansion |
Full mutation |
Patient’s Father |
11-Mar |
Normal |
Normal repeat |
|
Patient’s Mother |
10/pm |
Confirmed pre mutation |
Pre mutated allele |
|
Family 8 |
Patient |
16/nd |
Expansion |
Full mutation |
Patient’s Mother Patient’s |
16/pm |
Confirmed pre mutation Normal |
Pre mutated allele Normal repeat |
|
Patient’s Sister |
10-Mar |
Normal |
Normal repeat |
|
15-Nov |
||||
Family 9 |
Patient |
3/nd |
Expansion |
Full mutation |
Patient’s Father |
15-Sep |
Normal |
Normal repeat |
|
Patient’s Mother |
3/pm |
Confirmed pre mutation |
Pre mutated allele |
|
Family 10 |
Patient |
5/nd |
Expansion |
Full mutation |
|
Patient’s Father |
19-Sep |
Normal |
Normal repeat |
Patient’s Mother Patient’s |
5/pm |
Confirmed pre mutation Normal |
Pre mutated allele Normal repeat |
|
Patient’s Brother2nd |
9-May |
Normal |
Normal repeat |
|
Patient’s Sister |
11-Sep |
Normal |
Normal repeat |
|
11-May |
||||
Family 11 |
Patient |
3/nd |
Expansion |
Full mutation |
Patient’s Father |
11-Sep |
Normal |
Normal repeat |
|
Patient’s Mother Patient’s |
3/pm |
Confirmed pre mutation |
Pre mutated allele |
|
Patient’s Sister |
12-Sep |
Normal |
Normal repeat |
|
11-Mar |
Normal |
Normal repeat |
||
Family 12 |
Patient |
9/nd |
Expansion |
Full mutation |
Patient’s Father |
9/pm |
Confirmed pre mutation |
Pre mutated allele |
|
Patient’s Mother |
15-Mar |
Normal |
Normal repeat |
|
Family 13 |
Patient |
9/nd |
Expansion |
Full mutation |
Patient’s Father |
9/pm |
Confirmed pre mutation |
Pre mutated allele |
|
Patient’s Brother1st |
11-Mar |
Normal |
Normal repeat |
|
Patient’s Brother2nd |
11-Sep |
Normal |
Normal repeat |
|
Patient’s Sister |
16-Mar |
Normal |
Normal repeat |
|
Family 14 |
Patient |
5/nd |
Expansion |
Full mutation |
Patient’s Mother Patient’s |
5/pm |
Confirmed permutation |
Premutated allele |
|
Patient’s Sister |
12-Jun |
Normal |
Normal repeat |
|
15-Mar |
Normal |
Normal repeat |
||
Family 15 |
Patient |
9/nd |
Expansion |
Full mutation Normal repeat |
Patient’s Father |
10-Mar |
Normal |
Normal repeat |
|
Patient’s Sister |
15-Sep |
Normal |
||
Family 16 |
Patient |
9/nd |
Expansion |
Full mutation |
Patient’s Mother |
9/pm |
Confirmed permutation |
Premutation allele |
|
Patient’s Sister |
18-Dec |
Normal |
Normal repeat |
|
Patient’s Brother |
11-May |
Normal |
Normal repeat |
|
Family 17 |
Patient |
3/nd |
Expansion |
Full mutation |
Patient’s Father |
3/pm |
Confirmed pre mutation Normal |
Premutation allele |
|
Patient’s Mother Patient’s |
9-May |
Normal |
Normal repeat |
|
Patient’s Brother2nd |
20-Sep |
Normal |
Normal repeat |
|
Patient’s Sister1st |
11-Sep |
Normal |
Normal repeat |
|
Patient’s Sister2nd |
15-Mar |
Normal |
Normal repeat |
|
16-May |
Normal repeat |
|||
Family 18 |
Patient |
11/nd |
Expansion |
Full mutation |
Patient’s Father |
11-Mar |
Normal |
Normal repeat |
|
Patient’s Mother |
11/pm |
Confirmed pre mutation Normal |
Premutation allele |
|
Patient’s Sister |
18-May |
Normal repeat |
||
Family 19 |
Patient |
9/nd |
Expansion |
Full mutation |
Family 20 |
Patient |
10/nd |
Expansion |
Full mutation |
Patient’s Father |
10/pm |
Confirmed pre mutation Normal |
Premutation allele |
|
Patient’s Mother |
15-May |
Normal repeat |
||
Family 21 |
Patient |
5/nd |
Expansion |
Full mutation |
Table 2 Molecular profiling status of 21 DM1 families (n=77)
nd, Not detectable; pm, Premutation
CpG methylation had been done in all DM1 (molecularly confirm) and control samples. The DNA samples containing large CTG repeat expansion were methylated (Table 3) CpG neighbor to DMPK gene locus. Among, 21 positive samples, 13 samples were methylated and rest 8 (DM1 samples) & 50 (control samples) were unmethylated (Figure 3).
S.N. |
Disease status, mutation origin |
CTG repeat expansion |
Status of CpG methylation |
1 |
DM1,Unknown |
Large CTG expansion |
Methylated |
2 |
DM1,Paternal |
Small CTG expansion |
Unmethylated |
3 |
DM1, Paternal |
Large CTG expansion |
Methylated |
4 |
DM1,Maternal |
Large CTG expansion |
Methylated |
5 |
DM1, Unknown |
Small CTG expansion |
Unmethylated |
6 |
DM1, Paternal |
Small CTG expansion |
Unmethylated |
7 |
DM1, Maternal |
Small CTG expansion |
Unmethylated |
8 |
DM1, Maternal |
Large CTG expansion |
Methylated |
9 |
DM1, Maternal |
Large CTG expansion |
Methylated |
10 |
DM1, Maternal |
Large CTG expansion |
Methylated |
11 |
DM1, Maternal |
Large CTG expansion |
Methylated |
12 |
DM1, Paternal |
Small CTG expansion |
Unmethylated |
13 |
DM1, Paternal |
Large CTG expansion |
Methylated |
14 |
DM1, Maternal |
Large CTG expansion |
Methylated |
15 |
DM1, Unknown |
Large CTG expansion |
Methylated |
16 |
DM1, Maternal |
Small CTG expansion |
Unmethylated |
17 |
DM1, Paternal |
Large CTG expansion |
Methylated |
18 |
DM1, Maternal |
Small CTG expansion |
Unmethylated |
19 |
DM1, Unknown |
Small CTG expansion |
Unmethylated |
20 |
DM1, Paternal |
Large CTG expansion |
Methylated |
21 |
DM1, Unknown |
Large CTG expansion |
Methylated |
Table 3 Methylation status of the studied patients (n=21)
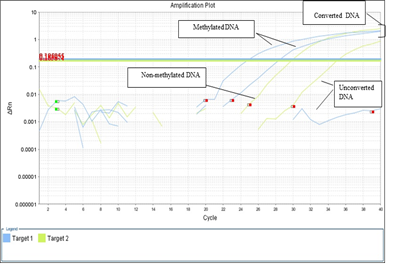
Figure 3 CpG methylation amplification plots of myotonic dystrophy and control samples. Blue (Methylated DNA), green (Non-Methylated), and clump lines show the converted DNA.
Correlation between CpG methylation with CTG repeat expansion, various clinical and biochemical parameters: CpG methylation was significantly correlated with CTG repeat expansion (r=0.679,p<0.001). However, it was not correlated with age at onset, hypersomina, aspiration, dysphasia, dyspepsia, and other parameters (Table 4).
Variables |
CpG methylation r(p-value) |
Variables |
CpG methylation r(p-value) |
Age at onset |
0.022(0.925) |
ADL |
-0.381(0.089) |
Age at presentation |
-0.159(0.491) |
Aspiration |
0.139(0.549) |
Duration |
-0.139(0.549) |
LWD |
-0.277(0.224) |
Facial weakness |
0.285(0.210) |
CTG repeat expansion |
0.679(<0.001) |
Dyspepsia |
0.010(0.967) |
CK |
0.552(0.069) |
Dysphasia |
0.347(0.124) |
Clinical Score |
0.062(0.789) |
Hypersomnia |
-0.062(0.789) |
MDRS |
0.037(0.872) |
Table 4 Correlation between CpG methylation and different parameters
LWD, Learning and writing disability
DM1 is a multisystemic autosomal dominant disorder and having phenomena of anticipation. The disabilities are substantial and therefore early detection is mandatory for reproductive counseling of families in which the DM1 has been observed as they have 50% chances of each pregnancy to have an affected child. The commonly used detection methods utilizes the combination of conventional PCR and triplet repeat-primed PCR. 24,33–36 The TP-PCR can detect the presence of long allele size without determining the total size of the expansion or the exact number of expanded CTG. Western countries routinely used TP-PCR while in India DM1 is mostly under clinical set up.
The MDSP-TPPCR method used as a convenient tool for the molecular diagnosis of DM1 in this study. The expansions were detected in all 21 patients while among 56 family members, except sixteen (permutated), 40 were normal. Apart from that, 50 normal individuals were subjected to molecular diagnosis of CTG repeat. A Correlation was observed between the incidence of disease and presence of CTG repeat alleles greater than 19 repeats in the normal population.31,37–39 The frequency of alleles with >19 CTG repeats differs according to ethnicity.7,40 The most frequent allele was different between Caucasians and Asians. Five repeats is the most common in European, and 12 repeats in Japanese and Koreans each.31,41,42 In our previous study, (in concordance with other studies),31,37,43,44 the frequency of CTG repeat >19 repeats was less than 6% in normal individuals and 2% in DM1 families normal individuals.8 Similarly, our 2018 study,8 highlighted the possible genotype-phenotype correlation between CTG repeat allele frequencies with different clinical, biochemical, molecular parameters and finding provide a clue about association of parameters with CTG repeat frequency.
In the present study, methylation status was studied in case (molecularly confirm)-control study and by using appropriate statistical analysis possible correlation had been observed between CpG methylation with CTG repeat expansion, clinical, biochemical, and laboratory findings. CpG methylation performed in all 21 DM1 (molecularly confirm) and control samples by use of bisulphate treatment and RT-PCR techniques. Among 21 positive samples, 13samples were methylated and remaining 8 (DM1 samples) & 50 (control samples) were unmethylated (Figure 3).
Our result highlighted that CpG sites adjacent to the CTG repeat expansion (large) were methylated, and this methylation was significantly correlated with CTG repeat expansion. However, it was not correlated with age at onset, hypersomina, aspiration, dysphasia, dyspepsia, and other parameters. These findings have same concordance with Shaw et al.,29 and Steinbach et al.,27 and contrary to Spits et al.,45 findings. Similarly in other triplet repeat disorder, level of methylation is positively correlated with the level of length expansion of the fragile X CCG repeat sequence.36 Though, the contribution of CpG methylation to disease pathogenesis is still not fully understood yet methylation might affect the expression of the toxic-CUG DMPK transcript, anti-sense-DMPK expression and/or Six-5 gene. CpG methylation also play an important role in the stability of repetitive sequences,25,26 and especially in mode of the CTG repeat & in the pathogenesis of disease.27,28
In summary, we had successfully established TP-PCR technique in the institute and it is a rapid and non-radioactive technique for detection of CTG repeat expansion in myotonic dystrophy and for other triplet repeat disorder like FRDA, FRAXA, SCA. TP-PCR is a substitute of southern blot (SA) because SA is radioactive and time consuming methodology. On other hand, status of CpG methylation was analyzed by bisulphite treatment, RT-PCR techniques. Next, its correlation with CTG repeat expansion, clinical, and biochemical parameters were studied and analyzed. However, more studies should be done in this direction to point out the involvement of CpG methylation in DM1 pathogenesis.
We are extremely grateful to all patients and their family members for their cooperation in this study and thankful to Sanjay Gandhi Post Graduate institute of Medical Sciences, Lucknow for providing infrastructure facility. Dr. Ashok Kumar is thankful to his Mentor Prof. Sarita Agarwal for time to time encouragement & guidance, Dr. Sunil Pradhan to providing samples and their clinical evaluation and to DST [NPDF/2015/000951] for providing fellowship..
The author declares that there are no conflicts of interest.
None.

©2020 Kumar, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.