Journal of
eISSN: 2373-4469


Research Article Volume 3 Issue 1
1Department of Genetics, Sanjay Gandhi Post Graduate Institute of Medical Sciences, India
2Department of Neurology, Sanjay Gandhi Post Graduate Institute of Medical Sciences, India
Correspondence: Department of Genetics, Sanjay Gandhi Post Graduate Institute of Medical Sciences, Lucknow, India, Tel 9152 2249 4349 (O), +9194 1533 6601, Fax 9152 2266 8017
Received: April 27, 2016 | Published: July 14, 2016
Citation: Kumar A, Agarwal S, Pradhan S. CTG repeat diversity at DMPK gene locus in Indian population. J Investig Genomics. 2016;3(1):7-13. DOI: 10.15406/jig.2016.03.00041
India has an extensive population admixture and genetic diversity of (CTG)n repeats. It is intriguing to investigate how different ethnic groups have a correlation with each other due to variability in (CTG)n repeats. Molecular evaluation of (CTG)n was performed on 400 individuals representing ethnic groups of North (Kayastha, Brahmin, Sunni, Shia, Vaishya, Chaturvedi) and North-East (Lachung and Mech) India. Population diversity analysis was made on the basis of phylogenetic, principal component analyses (PCA) and heat-map construction. Twenty three alleles (range 3-29 repeats) were detected when Indian subjects were pooled together. Kayastha, Brahmin, Sunni each showed 15 repeats as maximum percentage of common repeats, followed by Shia, Vaishya, Chaturvedi, Lachung and Mech respectively. Incidence for (CTG)>19 repeats was 5.87% in pooled group of India. In comparison to Indians (47.29%), higher frequency of (CTG)9-18 was concurred among Brazilians (100%) followed by North Americans (93.2%), Papua New Guineans (90%), Mexicans (86.2%) and Russians (83.4%). The North, North-East Indian populations form separate phylogenetic clades. However, Chaturvedis from mainland India formed a bridge between these two major population groups as evident from the dendrogram and genetic distance data. The PCA placed the North Indians, Caucasian and Mediterranean populations in one quartile and North-East Indian populations in the other. A bottleneck role of the north Eastern populations may be suggested as they serve as an inter-link between mainland India and Pacific/Australo-Malenesian populations from the human evolution perspective. In conclusion, our data provides an idea of (CTG)n repeat diversity in the studied Indian ethnic groups and their association with different ethnic and geographically isolated populations.
Keywords: (CTG)n repeat, north-east India, phylogenetics, population bottleneck
MEGA, molecular evolutionary genetic analysis;REML, restriction maximum likelihood; 6-FAM, fluorescein amidite/6-carboxyfluorescein; PCA, principal components analysis; DMPK, dystrophica myotonia protein kinase
The highly heterogeneous Indian population has been divided into more than two thousand ethnic groups mainly categorized into four races namely, Indo-Aryan (72%), Dravidian (25%), Mongoloid and others (3%). India lags only behind to Africa in possessing a vibrant linguistic, genetic and diverse cultural prototype1 The (CTG)n repeat diversity pattern is a sound approach to evaluate repeat expansions in human genome, including expansion mutations. Mostly trinucleotide repeat expansion disorders are distinguished with increased penetrance and disease severity2 in successive generations and parental sex bias in the transmission of the severe form of the disease which correlates with the degree of meiotic instability and allelic expansion. The rather frequent occurrence of triplet repeats in mRNA indicates that more loci containing unstable DNA expansions could be discovered.3 The most common clinical conditions featured with (CTG)n repeats are myotonic dystrophy (DM) and Spinocerebellar ataxia. DM has been distinguished on the basis of two different mutations into DM type 1 (DM1; Steinert disease) and DM type 2 (DM2; proximal myotonic myopathy or Ricker syndrome). DM1 being the most common form of muscular dystrophy in adults with an estimated incidence of 1:80004 is caused by a (CTG)n repeat expansion in the 3΄ un-translated region of the Dystrophica Myotonia Protein Kinase (DMPK) gene located within chromosome 19q13. The DMPK gene is of ~14kb length. It encodes 2.3kb of mRNA with 15 exons and the cAMP-dependent serine-threonine kinase protein comprising of 624 amino acids The (CTG)n repeats in DMPK gene vary in normal population with a range of (CTG)3-5 to (CTG)34. Severity of DM1 has been implicated with presence of (CTG)>50 repeats5‒8 DM2 is caused by a mutation in the ZNF9 (zinc finger protein 9) gene on chromosome 3q21. The first intron in ZNF9 contains a complex repeat motif (TG)n (TCTG)n(CCTG)n. Expansion of the CCTG repeat causes DM2.9,10 The repeat expansion for DM2 is much larger than for DM1, ranging from 75 to over 11000 repeats. Unlike DM1, the size of the repeated DNA expansion does not correlate with age of onset or disease severity in DM2. Anticipation is less evident clinically in DM2. A congenital form of DM2 has not been reported. Till date diversity of (CTG)n repeats have been studied mostly in disease populations. Previous studies have showed allelic distribution of the CTG polymorphism and the incidence of DM to be highly variable among various ethnic groups. (CTG)19-37 repeats have been associated with DM1 with an elevated prevalence among Brazilian and European11,12 populations followed by Korean13 Taiwanese14 and African15 populations. Similarly the incidence pattern for DM1 has also been observed at an increased percentage among Europeans16 than Africans17 Number of reports have suggested predisposition of (CTG)19-37 repeats towards DM1.12,18‒20 The present study has evaluated emphatically the impact of (CTG)n repeat diversity on variation among human races instead of disease severity. In the genetically kaleidoscopic Indian population, major studies need to be undertaken to unravel how ethnically diverse groups are associated with each other due to differential association of (CTG)n repeats. However, reports from NIMHANS (South India)19 Saha Institute of Nuclear Physics (East India)20 and Sanjay Gandhi Post Graduate Institute of Medical Sciences (North India) (Unpublished) showed lower incidence of (CTG)>19 repeats among highly diverse Indian population. We have designed the study in a manner to assess the importance of different (CTG)n repeats among thirty three ethnic groups/populations belonging to sixteen nations. This in turn will address whether the diversification in the (CTG)n repeats have played a role in human migration wave and in turn have an impact on altering the viral load.
Sample collection
2ml blood samples were collected in EDTA vials from 400 normal individuals from Northern and Northeast India. Informed consent was obtained from each subject. The study was performed after the approval of the institutional ethical reviewing committee of Sanjay Gandhi Post Graduate Institute of Medical Sciences (SGPGIMS), Lucknow. The mean age at sampling was 34.7±9.3. The blood samples collected from Northern India belongs to Kyastha, Brahmin, Sunni, Shia, Vaishya, Chaturvedi ethnic groups (p1=6), and Lachung and Mech tribes (p2=2) of Sikkim State (Northeast India). Blood samples were collected from 50 normal individuals from each group (p=p1+p2=8) (n=50x8=400).
Molecular genetic analysis
DNA extraction: Blood samples collected in EDTA vials were processed for DNA isolation by standard phenol chloroform method. The quality and purity of DNA was checked by measuring optical density (OD) at 260 nm to 280nm. The ratio of absorbance at 260 and 280 nm of DNA was around 1.7-1.9. The quality and purity was confirmed by 0.8% agarose gel electrophoresis in 1X TBE buffer and stored at -20°C till further use.
PCR and CTG repeat number analysis: The CTG repeat region in the DMPK gene was amplified by Myotonic Dystrophy Short PCR (MDSP) to determine the sizes of normal and/or permutated trinucleotide (CTG) repeats. PCR was performed in a reaction volume of 25µl using 50ng genomic DNA with 5 pmols of each primers 101-F (5’-FAM-CTT CCC AGG CCT GCA GTT TGC CCA TC-3’) and 102-R (5’-GAA CGG GGC TCG AAG GGT CCT TGT AGC-3’).5 FAM/6-FAM stands for fluorescein amidite/6-carboxyfluorescein (6-FAM) and it is commonly used fluorescent dye for labeling oligonucleotides. 6-FAM is reactive, water-soluble, and has an absorbance maximum of 492 nm and an emission maximum of 517nm. 6-FAM plays a particularly important role in real-time PCR applications, being used as a reporter moiety in TaqMan probes and Molecular Beacons. For such probes, 6-FAM is most commonly paired with the dark quencher BHQ-1, as the two have excellent spectral overlap. The cycling profiles were as follows: 5min at 95℃, 34 cycles of 10sec at 95℃, 30sec at 62℃, and 30sec at 72℃. A final extension at 72℃ for 10min completed the reaction. Repeat size was calculated by subtracting the number of base pairs of the flanking region from the total length of the PCR products and dividing the result by three. The fragment was analyzed by ABI PRISM 310 Genetic Analyzer with the Gene Mapper ID 3.1 software (Applied Biosystems, Foster City, CA, USA).
Data analysis and statistical methods: The frequency of (CTG)n repeat at DMPK locus for all 400 normal individuals were determined by direct counting. The observed incidence for specific CTG repeats among the studied populations was illustrated by Graph pad version 5.0. Phylogenetic analysis for common (CTG)n repeat frequencies was carried out by restriction maximum likelihood (REML) method using Molecular Evolutionary Genetic Analysis (MEGA), version 6.06 program (www.megasoftware.net). The principal components analysis (PCA) of DMPK gene frequencies were carried out using XLSTAT (Addinsoft®, France). Heat-map was plotted to illustrate the incidence of (CTG)9-18 repeat using the open heat-map tool (www.openheatmap.com). Frequency differences were illustrated via the default heat map colour gradient.
(CTG)n repeat distribution in Indian population and its comparison with other populations
The (CTG)n repeat length was determined in a total of 400 individuals belonging to eight Indian ethnic groups namely Kayastha, Brahmin, Sunni, Shia, Vaishya, Chaturvedi, Lachung, and Mech. Representative electropherograms showed patterns of (CTG)n repeat among Kayasthas (Supplementary Figure 1A) and Brahmins (Supplementary Figure 1B). Twenty-three alleles in range of 3-29 repeats were detected among the pooled samples and the percentage allele frequencies corresponding to (CTG)n repeats for each ethnic group were listed in Figure 1. The mean repeat size of pooled population was 4.0±6.74.
Eleven (CTG)n repeats were observed among Kayastha (58.33%), Brahmin (38%) and Sunni (31.16%). Meanwhile nine and four repeats were noted respectively for Shia (21.85%) and Vaishya (22%) groups. Interestingly three repeats each were found among the Chaturvedis (61.67%) and endogamous Lachung (63%) and Mech (71.67%) tribes (Figure 1) (Table 1). (CTG)>19 repeats were observed among Kayastha (6.22%), Brahmin (5%), Sunni (16.4%), Shia (11.34%) and Vaishya (8%) ethnic groups while Chaturvedi, Lechung, and Mech showed zero incidences (Table 1). The percentage frequency of (CTG)>19 repeat was 5.87% in the pooled group (Table 1) (Table 2). The majority of Indian ethnic groups showed two prominent peaks i.e.
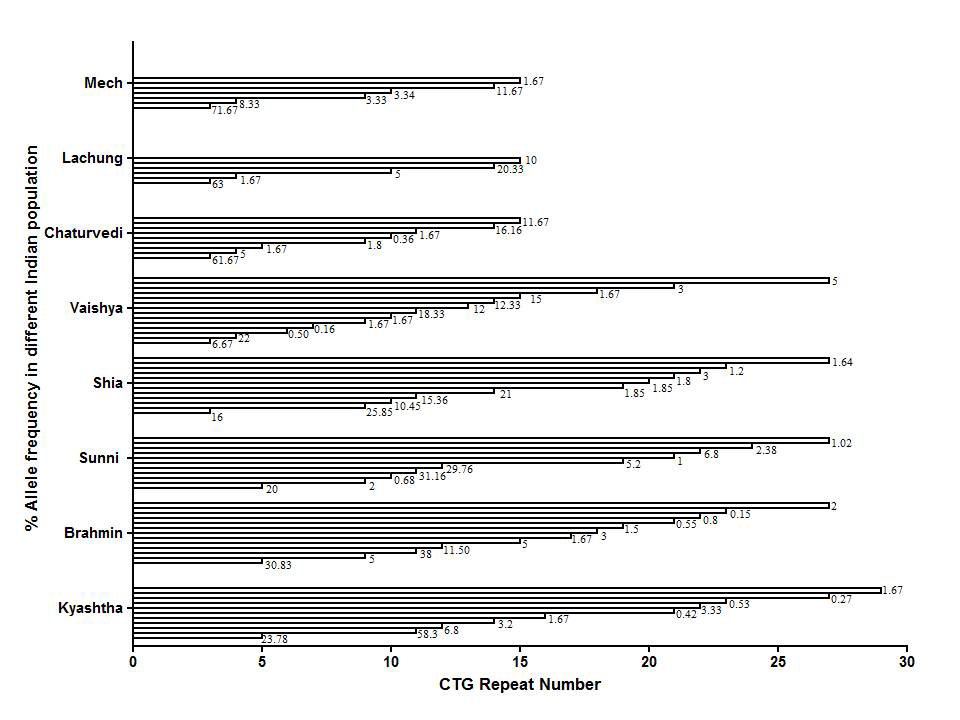
Figure 1 Dendogram showing the frequency of alleles (in %) from eight normal Indian population (p=8) subjects (neach =50).
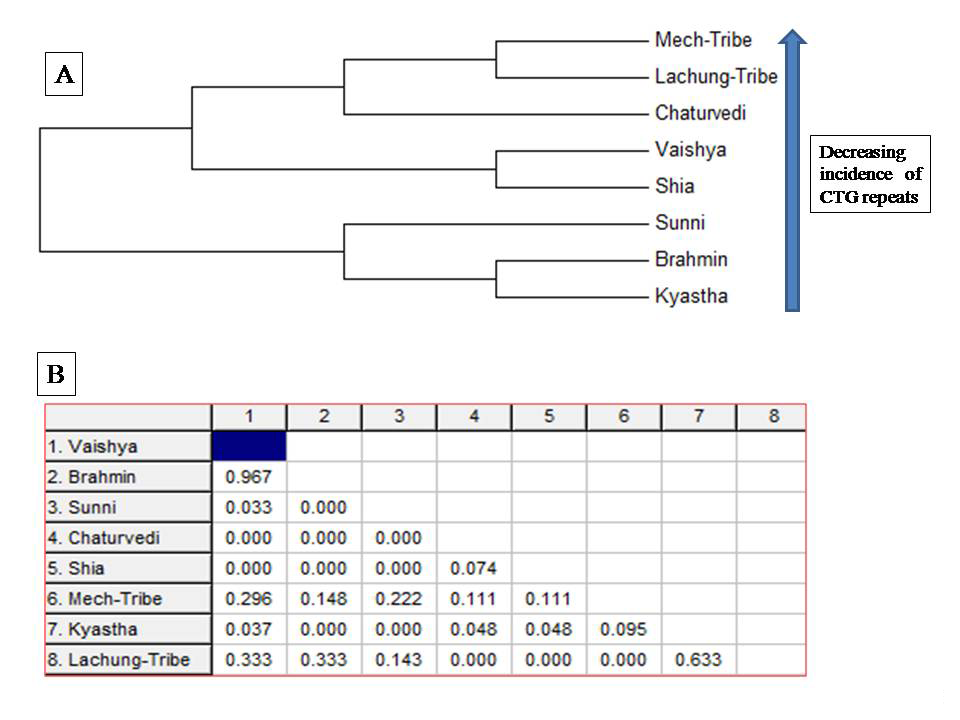
Figure 2 Phylogenetic analysis of some normal Indian population on the basis of % of maximum common CTG repeats.
No. of Samples in each Population |
Range of CTG Repeat Size |
No. of Alleles |
Repeat Size class and their Frequencies |
Percentage Frequency of Maximum Common CTG Repeat |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
Min |
Max |
=<5 |
6-8 |
9-18 |
≥19-30 |
|||||||
Kyastha (50) |
5 |
29 |
10 |
23.78 |
0 |
70 |
6.22 |
58.33% had 11 repeat |
||||
Brahmins (50) |
5 |
27 |
12 |
30.83 |
0 |
64.17 |
5 |
38% had 11 repeat |
||||
Sunni (50) |
5 |
27 |
10 |
20 |
2.68 |
60.92 |
16.4 |
31.16% had 11 repeat |
||||
Shia (50) |
3 |
27 |
11 |
16 |
26.3 |
46.36 |
11.34 |
25.85% had 9 repeat |
||||
Vaishya (50) |
3 |
27 |
13 |
28.67 |
4 |
59.33 |
8 |
22% had 4 repeat |
||||
Chaturvedi (50) |
3 |
15 |
8 |
68.34 |
1.99 |
29.67 |
0 |
61.67% had 3 repeat |
||||
Lachung (50) |
3 |
15 |
5 |
64.67 |
3.8 |
31.53 |
0 |
63% had 3 repeat |
||||
Mech (50) |
3 |
15 |
6 |
80 |
3.6 |
16.4 |
0 |
71.67% had 3 repeat |
||||
Pooled (400) |
3 |
29 |
23 |
41.53 |
5.31 |
47.29 |
5.87 |
26.28% had 3 repeat |
||||
Table 1 CTG repeat allele frequency in normal population of India
Population (2n), their Anthropological Affinity (no. Of chromosome) |
Range of CTG Repeat Size |
No. of Allele |
Percentage Frequency of (CTG)n |
||||
|---|---|---|---|---|---|---|---|
Size Class |
|||||||
Min |
Max |
=<5 |
8-Jun |
18-Sep |
19-35 (Size or Rangeb) |
||
Africana |
|||||||
Biaka (120) |
5 |
17 |
11 |
32.5 |
2.5 |
64.8 |
0.00 (NA) |
Mbuti (92) |
5 |
17 |
7 |
10.8 |
0 |
89.2 |
0.00 (NA) |
Bantu-speakers (358) |
5 |
22 |
13 |
24.9 |
7.3 |
67.3 |
0.50 (21-22) |
!Kung San (126) |
5 |
17 |
10 |
10.3 |
7.1 |
82.4 |
0.00 (NA) |
Ethiopian Jews (132) |
5 |
32 |
16 |
15.9 |
4.5 |
69.7 |
10.0 (20-32) |
European/Middle Easterna |
|
||||||
Yemenite jews(108) |
5 |
35 |
14 |
38.9 |
0 |
46.3 |
14.7 (20-35) |
Druze (100) |
5 |
24 |
11 |
42 |
0 |
54 |
4.00 (21-24) |
Danes (98) |
5 |
25 |
13 |
28.5 |
0 |
62.1 |
9.10 (21-25) |
Roman Jews (54) |
5 |
17 |
7 |
40.7 |
1.9 |
57.4 |
0.00 (NA) |
Mixed Europeans (78) |
5 |
29 |
12 |
37.2 |
1.3 |
56.3 |
5.20 (19-29) |
Micronesians (56) |
5 |
17 |
6 |
35.7 |
0 |
64.3 |
0.00 (NA) |
Nasioi (46) |
5 |
28 |
9 |
10.2 |
0 |
72.1 |
17.7 (21-28) |
New Guineans (40) |
9 |
21 |
9 |
0 |
0 |
90 |
10.0 (19-21) |
Pacific/Australo- Melanesiana |
|||||||
Micronesians (56) |
5 |
17 |
6 |
35.7 |
0 |
64.3 |
0.00 (NA) |
Nasioi (46) |
5 |
28 |
9 |
10.2 |
0 |
72.1 |
17.7 (21-28) |
New Guineans (40) |
9 |
21 |
9 |
0 |
0 |
90 |
10.0 (19-21) |
North Americana |
|||||||
Cheyenne (102) |
5 |
26 |
10 |
2.9 |
0 |
93.2 |
4.00 (22-26) |
Jemez Pueblo (86) |
5 |
27 |
9 |
8.2 |
0 |
87.3 |
4.7 (23-27) |
South Americana |
|||||||
Rondonian Surui (86) |
9 |
28 |
6 |
0 |
0 |
96.5 |
3.6 (21-28) |
Ticuna (122) |
9 |
18 |
5 |
0 |
0 |
100 |
0.00 (NA) |
Karitiana (102) |
9 |
17 |
3 |
0 |
0 |
100 |
0.00 (NA) |
Maya (102) |
5 |
28 |
8 |
9.9 |
0 |
86.2 |
4.0(22-28) |
Asian |
|||||||
Kochari (36)# |
5 |
17 |
5 |
25 |
0 |
75 |
0.00 (NA) |
Chinese (84)# |
5 |
17 |
7 |
36.9 |
0 |
63.1 |
0.00 (NA) |
Japanese (100)# |
5 |
28 |
11 |
19 |
0 |
72 |
9.00 (20-28) |
Yakut (102)# |
5 |
24 |
8 |
6.9 |
0 |
83.4 |
9.70 (21-24) |
Atayal (84)# |
5 |
17 |
6 |
45.3 |
0 |
54.7 |
0.00 (NA) |
Cambodians (48)# |
5 |
17 |
8 |
43.8 |
0 |
56.2 |
0.00 (NA) |
Indians (800) |
3 |
29 |
23 |
41.53 |
5.31 |
47.29 |
5.87 (19-29) |
Table 2 Comparison of percentage frequency of CTG Repeat Alleles in 26 Human populations including Indian population
Phylogenetic assessment of studied population
Based on the percentage of maximum common (CTG)n repeats frequency of the DMPK genes, phylogenetic analysis was carried out among eight Indian ethnic groups (Figure 2A). Populations were placed in a decreased orientation of (CTG)n repeats in the dendrogram. All the eight ethnic groups were divided into two main clusters. Kayastha, Brahmin and Sunni were observed in the first cluster while the second major cluster was found to be constituted of Shia, Vaishya, Chaturvedi, Lachung and Mech (Figure 2A). Upon drawing the phylogenetic assessment from (CTG)n repeat view point North-Indian ethnic groups like Kayastha, Brahmin and Sunni were found to be in close proximity followed by Shia, Vaishya and Chaturvedis. As expected the Mech and Lachung tribes were evidenced to be the least associated populations with north Indians in terms of (CTG)n repeat diversity (Figure 2A).
Genetic distance
The endogamous Mech and Lachung populations were in closest proximity (0.00) with each other. Interestingly two mainland Indian ethnic groups like Chaturvedi and Shia showed genetic nearness towards these two north eastern populations. North Indian populations like the Kayasthas revealed genetic distance wise association with Brahmins (0.00) and Sunnis (0.00) followed by Vaishya (0.037), Shia (0.048), Chaturvedi (0.048), and Mech (0.095) (Figure 2B).
Principal component analysis
Principal component analysis (PCA) was carried out in order to evaluate the association of the studied eight Indian ethnic groups with twenty five populations/ethic groups belonging to different countries (Figure 3). PCA findings complement the findings of phylogenetic analysis. The north Indian populations like Vaishya, Kayastha and Sunni showed a closer association with different ethnic groups belonging to Caucasian (Yemenite Jews, Danes, Yakut) populations, Asian (Yakut) population and with Pacific/Australo-Malaysians (Nasioi, New Guineans) as evident from the first quartile. Whereas the north Indian Brahmins showed nearness with mixed European and Druze populations. The north eastern Mech and Lechung tribes lie in close proximity with the Chaturvedis and were placed in the second quartile. Meanwhile north American populations (Cheyenne and Jemez Puebelo) along with South American populations (Maya and Radonian Surui) showed close association and as expected the Shia populations lie in proximal association with North African (Mubti and Kung San) populations (Figure 3). To a greater extent the results reflect geographic location wise robust association of the studied populations.
In this study we have evaluated the incidences of (CTG)n repeats at the DMPK locus among normal north and north-east Indian populations. The observation so drawn for (CTG)n repeat pattern among Indians has been compared there after with populations representing diverse ethnic and geographic origins. Further, analysis of (CTG)n repeats provided insights into how studied populations are interlinked with each-other and more importantly sheds light on the evolutionary origin of (CTG)n repeat. Few studies have been carried out studying the diversity of (CTG)n repeats with normal healthy populations. However, the (CTG)n repeats association with DM show a positive correlation between the disease incidence and frequency of normal (CTG)>19 repeats among Indian, European and Japanese populations.19‒23 Thus, it has been postulated that normal (CTG)≥19 repeats constitute a reservoir for recurrent expansion mutations.22,24 The results of our study show that there is considerable inter ethnic group variation in the frequencies of (CTG)n repeats. (CTG)3-5 repeat was found to be the smallest and most common allele in the majority of the ethnic groups while the combined frequency of (CTG)9-18 repeats was of maximum prevalence. These findings fell mostly in line with prior observations.12,18 The expansion of (CTG)n repeats into the large-sized range in humans appears to have originated in an ancestral northeastern-African population prior to the migration of modern humans out of Africa. This expansion of the (CTG)n repeat alleles may have crossed a threshold level of repeat number beyond which mutation occurs at a higher rate, explaining the broad distribution of (CTG)≥19 alleles outside Africa. Our data supports the model proposed by Imbert G22 which suggests, that the stability of (CTG)n repeat alleles is dependent on their length: accordingly (CTG)5 repeat alleles are most stable followed by (CTG)9–17 and (CTG)≥19.
Several studies have been reported on (CTG)n repeat variability.11,20,22,23,25,26 However, these were limited to single populations. We have studied the genetic diversity of (CTG)n repeats among thirty three ethnic groups and populations spread in sixteen countries. The maximum incidence of occurrence was found for (CTG)9-18 repeats cutting across the globe among different populations (Figure 4). Higher frequency of (CTG)9-18 repeat was concurred among Brazilian (100%) populations followed by populations from USA (93.2%), Papua New Guinea (90%), Mexico (86.2%) and Russia (83.4%). Pooled frequency calculated for eight Indian populations for (CTG)9-18 repeat stood at 47.29% (Figure 4). One of the most important factors which shape the population structure of a region is its position on the world map. Two major routes have been proposed for the initial peopling of East Asia; one through India to Southeast Asia and further to different regions of East Asia and the other via Central Asia to Northeast Asia, which subsequently expanded towards Southeast Asia and beyond.27 It is pertinent in this context that the Indian sub-continent has been considered as a major corridor for the migration of human populations to East Asia.28 Owing to its unique geographic position, Northeast India is the only region which currently forms a land bridge between the Indian sub-continent and Southeast Asia and acts as an important passage for initial peopling of East Asia. This may be the reason why we have found close genetic distance wise association among north eastern Mech and Lachung tribes with main land Indian ethnic groups like Chaturvedis and Shias who have travelled far from the Mediterranean to north India. In the case of India there exist an extensive population admixture and genetic heterogeneity. A recent finding has suggested that nearly all Indians carry genomic contributions from ancestral northern Indians related to central Asians, middle Easterners, western Eurasians and Europeans, while ancestral south Indians showed nearness to northern Indians.29 Largely endogamous and reproductively isolated groups are the pillars of societal pyramid in India. Hinduism; the most celebrated religion in the South Asian peninsular is based on a complex caste based system. Inter caste marriages are not a so common practice but marriages among sub-castes are allowed. So caste group present in same hierarchical cluster are observed to be biologically closer. On the contrary tribal groups don’t believe in any rudimentary caste based religious structure rather they rely on worshiping the nature and follow a strict endogamy.30 This may be the reason of less genetic diversity for (CTG)n repeats observed among the Lachung and Mech tribes.31 It signifies pre-dominance of lesser genetic diversity among the ancient tribes who are more prone to environmental imbalances in comparison to mainstream modern day populations.21,30,31 In conclusion, our data provides an idea of (CTG)n repeat diversity in the studied Indian ethnic groups and their association with different ethnic and geographically isolated populations.
We are indebted to all normal individuals for their cooperation in this study and thankful to Sanjay Gandhi Post Graduate institute of Medical Sciences (SGPGIMS), Lucknow for providing infrastructure facility. Ashok Kumar is thankful to DBT-New Delhi (DBT-JRF 2009-10/515) for awarding a Senior Research Fellowship.
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.

©2016 Kumar, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.