Journal of
eISSN: 2378-3184


Research Article Volume 8 Issue 6
National Institute of Oceanography and Fisheries, Egypt
Correspondence: Mohamed E Megahed, National Institute of Oceanography and Fisheries (NIOF), Gulfs of Suez & Aqaba’s Branch, Egypt
Received: December 05, 2019 | Published: December 16, 2019
Citation: Megahed ME. Genomic analysis of a decapod Penstylidensovirus 1 isolated from Penaeus semisulcatus. J Aquac Mar Biol. 2019;8(6):238-244. DOI: 10.15406/jamb.2019.08.00267
The purpose of this study was to identify the genomic characteristics of Penaeus stylirostris densovirus (PstDNV 1) affected farming of the green tiger shrimp Penaeus semisulcatus. Shrimp samples tested positive for PstDNV 1 by PCR and showing pathognomonic lesions were taken for cloning. Genomic DNA from infected shrimp was digested with restriction enzymes and plasmid libraries made before taking recombinant clones for shotgun sequencing. Altogether, 11 PstDNV1 contigs 3,530 to 3,912 nucleotides (nt) long were generated from P. semisulcatus. Each sequence contained ORFs encoding nonstructural (NS) proteins NS1 and NS2 and coat protein. The genome length of the isolates ranged from 3,598 to 3,912 bases. The GC content of the isolates varied from 42.5 % to 43.2%. These eleven PstDNV 1 isolates shared a pairwise identity of 96.4%. The phylogenetic analysis of the eleven PstDNV 1 isolates and the previously reported sequences showed the formation of 5 groups. Group 1 contains isolates from China, Brazil, California, Hawaii and 4 sequences from Egypt (KT316255.1, KT316260.1, KT316256.1 and KT316256.1). The second group contains sequences from Australia and 4 sequences from Egypt (KT316253.1, KT316252.1, KT316251.1 and KT316250.1). The third group clusters sequences from Thailand, Vietnam, Taiwan and two sequences from Egypt (KT316257.1 and KT316259.1). The appearance of PstDNV 1 in Egypt occurred after the introduction of Litopenaeus vannamei and Fenneropenaeus indicus. The complete genomes characterized in this study suggest that PstDNV was introduced to Egypt via movement of infected animals from distinct parts of the world. These finding serve as evidence that stricter monitoring protocols for the movement of shrimp species are fundamental as biosecurity measures to avoid the spread of pathogens that could potentially compromise the growth of the shrimp industry in Egypt.
Keywords: Penaeus stylirostrisDensovirus 1, PstDNV 1, IHHNV, Penaeus semisulcatus
Penaeus semisulcatus is the native shrimp species in Egypt cultured since the 1980s. With the introduction of Fenneropenaeus indicus, a nonnative species, shrimp farmers started experiencing disease outbreaks. Laboratory diagnosis revealed the presence of Penaeus stylirostris densovirus 1 (PstDNV1).
Penaeus stylirostris densovirus (PstDNV 1), commonly known as infectious hypodermal and a hematopoietic necrosis virus (IHHNV) is classified in the family Parvoviridae, sub-family Densovirinae.1 The virions are icosahedral, non-enveloped measuring 22-23nm in size, and contain single-stranded DNA of 4.1kb.2,3 The PstDNV 1 genome contains three major open reading frames (ORFs): left, middle, and right.2 The left ORF encodes a non-structural protein, NS1 containing 666 amino acids with a molecular mass of 75.77kDa similar to the NS1 protein of insect parvoviruses. The PstDNV 1, NS1 protein contains a conserved replication initiation motif, NTP-binding and helicase domain. The middle ORF completely overlaps with the left ORF, and encodes a protein of 363 amino acids (42.11kDa) with unknown function. The right ORF encodes the capsid protein containing 329 amino acids with a molecular mass of 37.5kDa.2 Transcription analysis has been carried out in PstDNV 1-infected P. vannamei shrimp.3,4 In the PstDNV 1 genome, in addition to three major ORF, there is a small ORF upstream of the left ORF (referred as NS1-a) that contains one putative 5’ donor site (D1). At the 5-end of the left ORF, there are three putative acceptor sites (A1, A2, and A3). The pre-mRNA representing the left ORF undergoes splicing at D1 position of NS1-a and A1 position of the left ORF transcript to generate a mature NS1 transcript.4 To date, the genome sequence of eight PstDNV 1 isolates are available in GenBank. These include two isolates from the Americas including an isolate from Hawaii (GenBank accession No. AF218266, 3909bp) and Mexico (AF273215; 3873bp), China, India, Korea, Vietnam, Taiwan, Thailand, Philippines and Indonesia.
PstDNV 1 was first reported in juveniles and sub-adults of blue shrimp (P. stylirostris) in super intensive raceways in Hawaii in early 80’s. The virus caused mass mortalities in P. styirostris.5 Later, the disease was found in Pacific white shrimp (L. vannamei) where it did not cause mortalities but resulted in growth retardation and deformities in appendages referred to as runt deformity syndrome (RDS).6 The virus is now endemic in the Americas, Asia and Australia.3
Shrimp aquaculture in Egypt has evolved from a subsistent farming activity in the early 1980s to an economically important industry by 2010. As penaeid shrimp farming becomes an important industry in Egypt, diseases have threatened the growth of this emerging sector of agriculture. Until 2010, P. semisulcatus was the native shrimp species that were cultured primarily in Egypt. In 2010, a non-native species, F. indicus was introduced. Following the introduction of F. indicus, shrimp farmers started experiencing disease outbreaks. The clinical signs in shrimp from affected farms indicated the presence of PstDNV 1. Histopathology and PCR diagnostic confirmed the presence of PstDNV 1 in shrimp displaying clinical signs. The complete genome of PstDNV 1 from samples originating in four farms was sequenced. This is the first report of PstDNV from a country in the Middle East. The importance of the detection of PstDNV 1 in shrimp aquaculture in Egypt is discussed as shrimp farming is becoming a major source of agriculture activities and revenue early for the country. The purpose of this study was to identify the genomic characteristics of Penaeus stylirostris densovirus (PstDNV 1) affected farming of the green tiger shrimp Penaeus semisulcatus in Egypt for possibility of application of effective control measure for this pathogen.
A total of 358 P. semisulcatus samples were collected from shrimp ponds at Dibah Triangle Zone (DTZ) in 2015 (Figure 1). The samples were preserved in 96% ethanol, and immediately stored in the laboratory. Total DNA and RNA were extracted from pleopods and hepatopancreas of shrimp using DNeasy and RNeasy minikits (Qiagen) following manufacturer’s protocols. DNA from each specimen was then amplified via PCR using the primer pair IHHNV648F (GAACGGCTTTCGTATTTTGG), and IHHNV648R (AGCGTAGGACTTGCCGATTA).7 The reaction mixture contained Master Mix 12.5μl, Primer mix 2.5μl, Q-solution 2.5μl, and 2.0μl total genomic DNA in a total reaction volume of 25μl. The thermal profile for the PCR amplification included an initial denaturation at 950C for 5 minutes, followed by 35 cycles at 940C for 30 seconds, annealing at 590C for 1 minute 30 seconds, extension at 720C for 1 minute 30 seconds, and an extension at 720C for 10 minutes. The PCR amplicons were electrophoresed using a 1.5% agarose gel, stained with ethidium bromide, and photographed (Safe Blue Illuminator/ Electrophoresis System, MBE-150-PLUS, MajorScience, USA).
Histopathology
Histological processing was performed following published protocol.5 Briefly, animals displaying clinical signs of PstDNV 1 infection were fixed in Davidson fixative. Tissue sections of Davidson-fixed were stained with hematoxylin and eosin, and examined under a microscope (Model name of the microscope and vendor’s name).6-8 The diagnostic procedure included the examination (at 200X and 400 X bright fields) of the stained sections of the gills, pleopod, maxilliped, stomach mucosa, ventral nerve cord, ganglia, hematopoietic tissues and antennal gland. Diagnostics was considered positive for PstDNV when Cowdry type A inclusions bodies were observed in hypertrophic nuclei of cells within any tissue of the tissues or organs listed above.5
PstDNV 1 genome sequencing and sequence analysis
Shrimp samples tested positive for PstDNV 1 by PCR and showing pathognomonic lesions were taken for cloning. Genomic DNA from infected shrimp was digested with restriction enzymes and plasmid libraries made before taking recombinant clones for shotgun sequencing using an ABI 3730XLS sequencer (Macrogen Inc., Seoul, South Korea). Contigs were generated and aligned to the viral reference genomes (accession number Hawaii isolate: AF218266) to separate viral contigs from shrimp genome contigs. Sequences were examined using Chromas Version 1.459 and BioEdit 7.0.9, open reading frames (ORFs) were annotated using BLASTn, Geneious R11 (Biomatters) and multiple alignments were performed using CLUSTALW.
The nucleotide and protein sequences were analyzed using Geneious R11 (Biomatters). All together 11 PstDNV 1 contigs were obtained. The ORFs in eleven isolates were predicted using GeneMark10 and Geneious R11. The annotation and the identity search of the ORFs were conducted by BLASTx and Geneious. The pairwise identity (nucleotide and protein) of the Egypt isolates with the Hawaii reference strain (AF218266.2) was calculated by CLUSTAL W within the program Geneious R11 and BLASTx. The nucleotide sequences of the 11 IHHVN isolates were also aligned by CLUSTAL W within the program Geneious R11. A list of PstDNV 1 isolates were used for the phylogenetic analysis (Table 1).
Region |
Gen Bank Accession number |
Date of publication |
Hawaii (Reference strain) |
AF218266.2 |
2009 |
California |
AF273215.1 |
2014 |
Australia |
KM593908.1 |
2015 |
Australia |
KM593909.1 |
2015 |
Australia |
KM593910.1 |
2015 |
Australia |
KM593911.1 |
2015 |
Thailand |
AY362547.1 |
2004 |
Vietnam |
KC513422.1 |
2013 |
Taiwan |
AY355307.1 |
2004 |
Thailand |
AY102034.1 |
2003 |
India |
GQ411199.1 |
2014 |
Tanzania |
AY124937.1 |
2014 |
Brazil |
KJ862253.1 |
2014 |
China |
JX258653.1 |
2012 |
Table 1 A list of PstDNV 1 isolates used for the phylogenetic analysis
To assess the phylogenetic relation and to elucidate the possible origin of the 11 IHHNV isolates from Egypt, the genome sequences of IHHNV isolates from other regions of the world were obtained from GenBank for phylogenetic analysis (Table 2). The sequences were aligned by CLUSTAL W within the program Geneious R11. The phylogenetic tree was constructed by using the Maximum Likelihood method.11 The percentage of trees in which the associated taxa clustered together is shown next to the branches. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using the Maximum Composite Likelihood (MCL) approach, and then selecting the topology with superior log likelihood value. The complete VP (viral capsid protein) sequence (990nts) was used for the phylogenetic analysis. A total of 25 nucleotide sequences were utilized. Evolutionary analyses were conducted in MEGA7.12
Isolate |
Host |
Sample location |
GenBank Accession No. |
Genome length |
%GC |
%nt Identity |
%NS1-b identity |
%NS2 identity |
%VP identity |
Decapod |
Penaeus semisulcatus |
Egypt (Alexandria Abu |
KT316250.1 |
3,610 |
42.7 |
97.29 |
98.05 |
97.25 |
98.18 |
Decapod penstyldensovirus 1 |
Penaeus semisulcatus |
Egypt (Idku, 31° 19' |
KT316251.1 |
3,749 |
42.9 |
97.84 |
98.65 |
98.35 |
97.88 |
Decapod penstyldensovirus 1 |
Penaeus semisulcatus |
Egypt (Damietta, 31° |
KT316252.1 |
3,749 |
42.9 |
97.95 |
98.95 |
98.63 |
97.88 |
Decapod penstyldensovirus 1 |
Penaeus semisulcatus |
Egypt (PortSaid, 31° |
KT316253.1 |
3,598 |
42.7 |
98.69 |
99.40 |
99.45 |
99.09 |
Decapod penstyldensovirus |
Penaeus semisulcatus |
Egypt (Arish, 31° |
KT316254.1 |
3,824 |
43.0 |
98.64 |
98.80 |
99.18 |
98.79 |
Decapod penstyldensovirus 1 |
Penaeus semisulcatus |
Egypt (Dibah Triangle Zone, |
KT316255.1 |
3,901 |
42.8 |
97.95 |
99.55 |
99.45 |
99.70 |
Decapod penstyldensovirus 1 |
Penaeus semisulcatus |
Egypt (Gulf of Suez, |
KT316256.1 |
3,909 |
43.0 |
100 |
100 |
100 |
100.00 |
Decapod penstyldensovirus 1 |
Penaeus semisulcatus |
Egypt (Lake Bardawil, |
KT316257.1 |
3,824 |
43.2 |
95.79 |
96.40 |
96.43 |
97.27 |
Decapod penstyldensovirus 1 |
Penaeus semisulcatus |
Egypt (Burullus, 31° |
KT316258.1 |
3,908 |
42.9 |
93.77 |
91.89 |
95.88 |
96.36 |
Decapod penstyldensovirus 1 |
Marsupenaeus japonicus |
Egypt (Lake Bardawil, |
KT316259.1 |
3,912 |
43.2 |
95.60 |
95.80 |
95.88 |
97.27 |
Decapod penstyldensovirus 1 |
Marsupenaeus japonicus |
Egypt |
KT316260.1 |
3,837 |
43.0 |
99.12 |
99.55 |
100 |
99.09 |
Table 2 General information from the 11 IHHNV isolates from Egypt. The table shows general information of each strain such as: isolate name, host, sample location and GenBank accession number. Also shown are the genome lengths, GC content (%GC), % of pair wise nt (%nt identity) and % of pair wise identity of the three major ORFs (% NS1-b identity, % NS2 identity and % VP identity) when compared to reference strain AF218266.2
PstDNV 1 caused retardation in growth and smaller size at harvest in juveniles and adults of P. semisulcatus. Infected shrimp with acute infection showed reduction in food consumption became lethargic and were often cannibalized by healthier shrimp. Infected P. semisulcatus presented a mottled appearance. Histopathology of PstDNV 1in P. semisulcatus is shown in H&E stained sections with severe acute intranuclear, haloed, eosinophilic Cowdry type A (CAI) inclusion bodies were evident in some infected cells. PstDNV1-infected cell nuclei. Hematopoietic nodule, and cytoplasmic accumulations of PstDNV 1 are apparent in developing oocytes in the ovary where IHHNV appears to be being deposited as yolk (Figure 2).
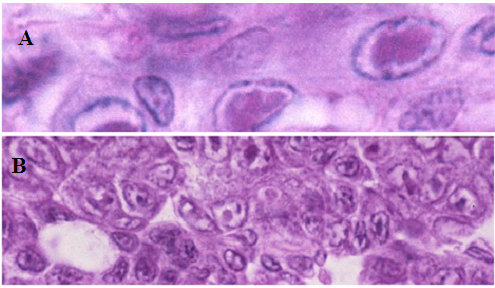
Figure 2 Histopathology of Penaeus semisulcatus showing eosinophilic Cowdry A type lesions in (A) gills and (B) hematopoietic nodule.
PCR detection of PstDNV 1 in semisulcatus
Penaeus semisulcatus displaying clinical signs and pathognomonic lesions of PstDNV-1 was tested by PCR using the primer pair IHHNV648F/IHHNV648R primer set (Figure 3).
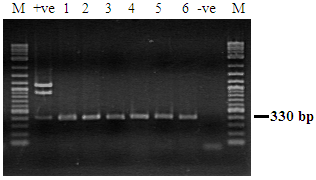
Figure 3 Detection of PstDNV 1 in Penaeus semisulcatus by PCR. Lane M: Molecular weight marker (100 bp). Lane +ve: positive control; Lanes 1-6: IHHNV infected samples. Lane -ve: IHHNV negative samples. Lane M: Molecular weight marker (100 bp).
Analyses of PstDNV 1 genome sequence
PstDNV1 and HPV are classified in the family Parvoviridae, genus Brevidensovirus.13Altogether, 11 PstDNV1 contigs 3,530 to 3,912 nucleotides (nt) long were generated. Each sequence contained ORFs encoding nonstructural (NS) proteins NS1 and NS2 and coat protein, as reported for other PstDNV1 isolates.2,3 A pairwise comparison of PstDNV1-Egypt isolates to the reference strain (GenBank accession number AF218266)14 showed 92.33% to 100% identity.
Altogether eleven unique contigs were generated for PstDNV 1 from P. semisulcatus. The genome length of the isolates ranged from 3,598 to 3,912 bases (Table 2). The GC content of the isolates varied from 42.5 % to 43.2%. These eleven PstDNV 1 isolates shared a pairwise identity of 96.4%. The three major ORFs, NS1-b, NS2 and VP were identified in all isolates. The NS1-b ORF presented a length of 2,154 bases in all isolates with the exception of isolate KT316255.1 (2,001 bases). Upstream to NS1-b a small ORF, NS1-a, was identified in most isolates with the exception of isolates KT316256.1 and KT316258.1. In KT316256.1 the NS1-a ORFs was annotated as two ORFs (NS1-a1 and NS1-a2) while in KT316258.1 this ORF was not identified (Figure 4).
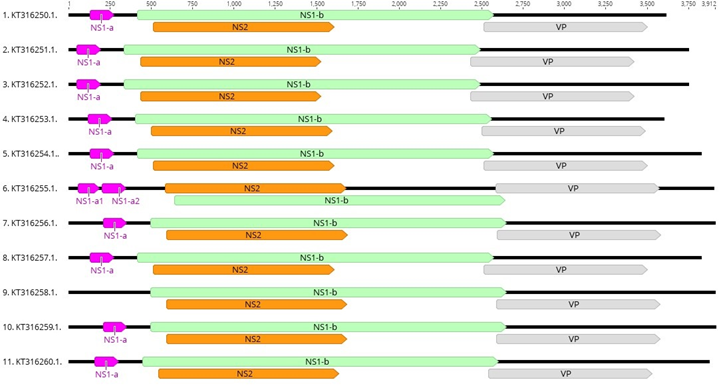
Figure 4 Schematic representation of the genome organization of eleven PstDNV 1 isolates from Egypt. The genome of PstDNV 1-Egypt isolates contains three major ORFS: NS1-b (Green), NS2 (Orange) and VP (grey). Upstream of NS-1b, a small ORF, NS1-a (pink) is shown. All isolates with the exception of KT316255.1 and KT316258.1 share the same ORFs. The isolate KT326255.1 has two ORFs (NS1-a1 and NS1-a2) that are homologous to NS1-a. Additionally, isolate KT316268 is missing ORF, NS1-a.
Differences were identified in the replication Motifs I and II in the NS1-b sequences (Figure 5). In isolate KT316255.1 substitution S226C was detected in Motif I. In Motif II substitutions I358 and L359F were observed in KT316254.1. In the conserved NTP-binding and helicase domains A, B, and C only sequence KT316254.1 shows an aa substitution T627I in domain “C” (Figure 6).

Figure 5 Replication initiator motifs I and II in NS1-b protein from the PstDNV 1 isolates from Egypt. Motif I and Motif II extend from positions 309-366 in all isolates with the exception of isolate KT316255.1 where Motif I and Motif II extend from positions 258-315. Only isolate KT316255.1 shows an aa substitution (S226C) in Motif I. Two aa substitutions I358L and L359F were observed in Motif II in sequence KT316254.1. The consensus sequence is shown on the top and the blue arrows indicate the Motif sequences. Amino acids substitutions are marked in white.

Figure 6 A conserved region of 119-aa of the NS1-B sequence, NTP-binding and helicase domains A, B, and C in the PstDNV 1 isolates from Egypt. The 119-aa sequence extends from positions 523-641 in all isolates, with the exception of sequence KT316255.1 where this sequences extends from positions 472-590. Only sequence KT316254.1 shows an aa substitution T627I in domain “C”. In the rest of the isolates all domains have identical sequences. The consensus sequence is shown on the top and the blue arrows indicate the NTP-binding and helicase domains A, B and C. Amino acids substitutions are marked in white.
Phylogenetic analysis of PstDNV 1 isolates from Egypt
The phylogenetic analysis of the eleven PstDNV 1 isolates and the previously reported sequences showed the formation of 5 groups. Group 1 contains isolates from China, Brazil, California, Hawaii and 4 sequences from Egypt (KT316255.1, KT316260.1, KT316256.1 and KT316256.1). The second group contains sequences from Australia and 4 sequences from Egypt (KT316253.1, KT316252.1, KT316251.1 and KT316250.1). The third group clusters sequences from Thailand, Vietnam, Taiwan and two sequences from Egypt (KT316257.1 and KT316259.1). Group 4 contains an isolate from India and Egypt (KT316258.1). Finally, group 5 is represented by a single sample Tanzania (East Africa) (Figure 7). These results suggest that the IHHNV strains in Egypt might have been imported from distinct regions of the world.
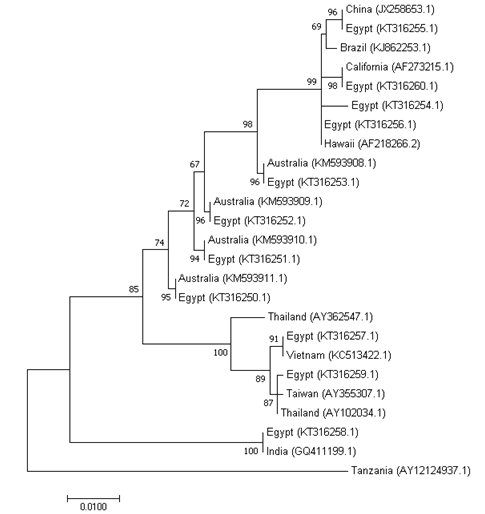
Figure 7 Maximum likelihood tree obtained from the nucleotide sequence of the complete capsid protein gene of PstDNV 1 isolates. The Evolutionary relationships of the eleven PstDNV 1 isolates from Egypt and twelve isolates from other geographic regions are presented. The percentage of trees in which the associated taxa clustered together is shown next to the branches. The tree is drawn to scale and the branch length was measured in the number of substitutions per site. GenBank accession number of different PstDNV 1 isolates is given in parentheses.
The present study provides important information regarding the sustainability of shrimp culture in Egypt. Even though genetically improved shrimp pathogen free (SPF) stocks are being imported and currently used for shrimp culture practice in Egypt, these are still susceptible to the viruses that cause mortalities and loss to shrimp farmers. The shrimp Penaeus semisulcatus at DTZ infected with IHHNV show slow growth rate, deformity, and low survival rate, a clinical signs specific to IHHNV infection. Due to the infection with IHHNV, farmers start harvesting shrimp at an early culture period (maximum 60 days) to avoid crop loss. The mortality of shrimp due to IHHNV infection has been reported in P. stylirostris,5 and heavy mortalities of P. monodon in India was reported due to IHHNV infection.15
In Egypt, wild-caught P. semisulcatus broodstock are the only source of spawners in hatcheries. Screening of broodstock for viral infections is not a routine practice in Egyptian hatcheries, potentially allowing viral transmission and the contamination of aquaculture facilities. Thus, the high prevalence of IHHNV/PstDV-1 infection in all hatcheries and grow-out ponds could be due to using wild P. semisulcatus broodstock naturally infected with the virus or to other wild crustaceans entering the shrimp ponds during culture. Therefore, the high prevalence of PstDV-1 in juvenile shrimp in grow-out ponds could be a consequence of low awareness regarding viral screening in hatcheries, meaning that shrimp seed released for stocking would carry a high potential risk of being infected by multiple viral infections contracted in the hatchery.
The NS1-b ORF presented a length of 2,154 bases in all isolates with the exception of isolate KT316255.1 (2,001 bases). In KT316256.1 the NS1-a ORFs was annotated as two ORFs (NS1-a1 and NS1-a2) while in KT316258.1 this ORF was not identified. The well known geographic ranges of infectious PstDV-1 has been reported from the Americas, Asia and Southeast Asia, Australia, Middle East and many Pacific islands,16 this is in line with the results obtained in the present study. Phylogenetic analysis showed that the eleven PstDNV 1 isolates and the previously reported sequences showed the formation of 5 groups.
This study showed that the infectious PstDV- 1 strain found had a closer similarity with the China and Brazil strains. The high level infections by IHHNV have high impact on P. semisulcatus growth and reproductive performance. The existence of PstDV-1 in Egyptian aquaculture has negative impact of the future of shrimp culture in Egypt. The primer pair IHHNV648F/ IHHNV648R primer is appropriate for amplifying the PstDV-1 in Egypt, and therefore it is suitable to use for routine PstDV-1 diagnosis of shrimp broodstock and postlarvae before spawning in the hatchery and farm stocking. Shrimp hatcheries have experienced difficulties in obtaining high quality broodstock and seeds of P. semisulcatus. Recently, shrimp farmers in Egypt have switched from P. semisulcatus to L. vannamei due to the availability of SPF of high quality broodstock and seed. However, in some farmers have remained prefer the use of P. semisulcatus although cultivation of L. vannamei has begun since 2015. Future studies are suggested to continue identify major pathogens affected shrimp farming in Egypt and establish a biosecurity measures and breeding program for selection of disease resistant shrimp strains.
The emergence of several pathogen of viral origin has had a serious economic impact on shrimp aquaculture in Egypt since 2010 with the first introduction of F.indicus and has severely threatened the sustainability and growth of shrimp farming practice. In Egypt cultured shrimp P.semisulcatus has been found exhibiting the typical signs of PstDV-1 and this condition has greatly affected production in a negative way. These finding serve as evidence that stricter monitoring protocols for the movement of shrimp species are fundamental as biosecurity measures to avoid the spread of pathogens that could potentially compromise the growth of the shrimp industry in Egypt. Therefore, strict biosecurity measures are necessary to prevent the shrimp from such kinds of infection. As white shrimp L. vannamei culture practice is just picking up, it is essential to adopt stringent measures to make shrimp farming sustainable.
GenBank Submission: The nucleotide sequence of Penaeus stylirostris Densovirus 1 (PstDNV1) from Penaeus semisulcatus reported in this paper have been submitted to the GenBank with the accession numbers: KT316249.1, KT316250.1, KT316251.1, KT316252.1, KT316253.1, KT316254.1, KT316256.1, KT316257.1, KT316258.1, KT316259.1 and KT316260.1.
The authors would like to the farmers at DTZ, Egypt for providing shrimp samples and information regarding the history of the disease outbreaks. This work was supported by Science & Technology Development Fund grants 5661.
The author declares that there are no conflicts of interest.

©2019 Megahed. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.