
Research Article Volume 4 Issue 5
Bayesian Estimation of the Number of Individuals in a Sample with a Known Weight
Samu M,1
Regret for the inconvenience: we are taking measures to prevent fraudulent form submissions by extractors and page crawlers. Please type the correct Captcha word to see email ID.

Atso Romakkaniemi,2 Elja Arjas3
1Department of Mathematics and Statistics, University of Helsinki, Finland
2Natural Resources Institute Finland
3Department of Mathematics and Statistics, University of Helsinki, Finland
Correspondence: Samu Mäntyniemi, Department of Environmental Sciences, University of Helsinki, Finland
Received: October 27, 2016 | Published: November 15, 2016
Citation: DOI: 10.15406/jamb.2016.04.00095
Download PDF
Abstract
We introduce a Bayesian probability model for making inferences about the unknown number of individuals in a sample, based on known sample weight and on information provided by subsamples with known weights and corresponding counts. Inherent in the Bayesian approach, the model allows for an incorporation of prior information that is often available about the sample size and other uncertain parameter values. As a result, the model provides an estimate of the number of individuals in the sample in the form of a posterior probability distribution that includes both the prior information and the interpretation of the observed data. Such a result cannot be obtained using the frequentist approach. The model presented here can be applied to a wide range of similar problems. Here our main focus is stock assessment, where the task is the conversion of the catch weight into the number of individuals in the catch. The model is easy to use due to availability of general purpose MCMC simulation software, and it can be used either in a standalone fashion or embedded into more complex probability models.
Keywords:Catch samples, Markov chain monte carlo (MCMC), Sample size, Weight distribution
Introduction
Counting the number of individuals in a large sample can be very laborious or impractical. Instead of exact counting of all individuals, the size of a large sample can be estimated by weighing it and then using information about the mean weight of individuals obtained from smaller samples. Estimation approaches based on this general idea have a number of natural applications in aquatic sciences. For instance, the number of fish in a commercial catch is often estimated in this way in order to obtain data that would be suitable for typical stock assessment methods.1,2 Further, the approach is apparently commonly used in the estimation of the number of fish raised in fish hatcheries for subsequent stocking.3 Besides estimating the number of fish, the approach has been applied in e.g., estimating the number of eggs in fish gonads.4 However, estimates obtained this way will always involve an element of uncertainty unless all individuals in the sampled population are of exactly equal weight, and unless measurement error is negligible. For a systematic analysis, this uncertainty should be taken into account in all further considerations utilizing these estimates. In fisheries science such uncertainty has been often neglected, or frequentist methods have been applied. Unfortunately frequentist methods cannot provide measurements of uncertainty about parameters, even though the results of the frequentist analysis are often phrased in a way which invites such a misinterpretation.
The Bayesian approach to statistical inference provides a flexible framework for working with multiple levels of uncertainty, and is therefore becoming increasingly popular in fisheries science. In the Bayesian approach, uncertainty is described by assigning probability distributions to quantities whose values are uncertain. Thus, in particular, uncertain values of the sample size are presented in terms of corresponding probability distributions. This paper presents the development of a Bayesian probability model which can be used to derive probabilistic estimates of the size of a sample of a known weight. The model structure is introduced by considering samples of fish, but the same logic applies to any kind of comparable items like invertebrates, plants, stones etc.
Sampling
A sample containing N fish can be obtained from a fish population either by simple random sampling or by some form of selective sampling. In a typical case N is large, but the theory presented here holds in principle for any positive integer value. On the other hand, the population from which the sample is drawn is assumed to be infinitely large. For example, fish living in a particular rearing pond are seen as a sample from the potentially infinitely large population of similar fish that could be produced in the given pond and under conditions similar to present ones.
The sample of size N is then divided without selection into k +1 subsamples, of which one is typically large compared to the others. Here we denote the number of fish in the large sub sample by n* and the number of fish in the smaller Sub samples by nj, j = 1,…,k, assuming then that
The weight (s*) of the large subsample and the weights (sj) of the smaller subsamples are assumed to have been measured accurately enough to be treated as known, as well as the number of fish from corresponding counts of the smaller subsamples. As a consequence, only n* remains unknown and needs to be estimated. If sampling from the fish population is made without any form of size selection, samples of sizes n*, n1,….,nk can be obtained as independent samples directly from the population, and in any order.
Probability model
We begin by making the assumption that the weights (wi; i=1,…,N) of individual fish are exchangeable for all values of N. This assumption means in particular that the joint predictive distribution of the weights, describing beliefs about the weights of the fish in the sample, is always the same regardless which particular N fish would have been sampled, and how they would be ordered within the sample. According to the celebrated representation theorem of de Finetti, the assumption of exchangeability allows us to write the joint predictive distribution (density) in the form
(1)
Where f is an unknown density function, and Q(f) denotes a probability measure over all distribution functions. This can be interpreted as if we had N independent fish weights taken from an unknown weight distribution function f, which again is assigned a prior probability distribution Q. The operational interpretation of Q (f) is then ”what we believe the empirical weight distribution would look like for a large sample”.5 Our second assumption is a convention which we make to simplify the analysis: we restrict the set of possible weight distribution functions to parametric distribution families F, for which it holds that if the individual weights wi follow independently a fixed distribution f belonging to distribution family F, then also arbitrary finite sums
follow a distribution which belongs to the same distribution family. Gamma and Normal families are known to satisfy this condition, and from now on we assume that F is either of these two families. Within the chosen family (Gamma or Normal), prior uncertainty about the weight distribution can be expressed by assigning a prior distribution to two parameters of the distribution family. Here we use mean μ and standard deviation σ , but other parameterizations can be used as well. Then the joint predictive distribution of the individual weights can be written as
(2)
where
is the Gamma or Normal density, and
is a joint prior distribution of its parameters. Because individual weights are conditionally independent given μ and , the conditional expected value and conditional standard deviation of the sample weight are
and
. This implies that the joint predictive distribution for the sample weights, given the sample sizes n1,….,nk can be written in the form
(3)
This model can be also specified by the following sequence of definitions:
(4)
where D(mean; standard deviation) in each case denotes a suitable prior probability distribution. The Normal distribution can be used for sample weights if all nj:s are large, as the distribution of the sum of independent random variables approaches the Normal distribution when nj increases, regardless of the shape of the distribution of individual weights. It should be noted, however, that the Normal distribution allows also for negative weights, which is not realistic. Prior distributions for μ,σ2 and n* can have any shapes, as long as it is recognized that they can have only positive values. If the sample weights (s1,…sk, s* ) are not observed without non-negligible error, the model can be extended to account for measurement error by treating the true sample weights as unknown and by adding an extra layer to the model specification:
(5)
where observed weight measurements (m1…mk, m* ) and corresponding standard deviations (v1…vk, v*) are assumed to be known. The form of the measurement error distribution can be in principle chosen in any way that would seem appropriate in the given context. Information about the shape of the distribution and about its standard deviation could come from expert judgement and/or from an independent study of the measurement error. Despite the simple model structure, the posterior distribution is analytically intractable and approximation methods are needed for a numerical evaluation of the probabilities of interest. Our approach is to use Markov chain Monte Carlo (MCMC) simulation 6 to draw a large number of samples from the posterior distribution, and use corresponding sample averages as summaries of the posterior distribution. This task can be accomplished easily by using a general purpose MCMC software, like Win BUGS 7.
Example: Number of fish in a rearing pond
Suppose that all N fish were captured from the rearing pond and moved to a tank. We begin by making the assumption that the weights wi of the fish in the rearing pond are a conditionally independent sample from a Gamma distribution characterized by unknown mean and coefficient of variation
. The sample of size N was then divided into four subsamples, of which one has unknown size n*, and the others are of known sizes n1=108, n2=101, n3=115. It is also assumed that the manufacturer of the scale has specified that the observed weights vary symmetrically around the true weight with standard deviation of 10g. Here we use a Normal distribution to describe the variation of the measurements (mj, m*) around the true value. Prior distributions for model parameters N, and were obtained by interviewing an expert who is familiar with local aquacultural practices. He was told that the rearing pond had bottom area of 50m2, it was located in Northern Finland and contained two-year-old salmon smolts. We formalised his prior beliefs by the following prior distributions:
(6)
In addition to the above specification, parameter u was constrained to lie in the interval [0,20 000] and parameter in the interval [5,60], that is, within these intervals the prior probability density is proportional to the distributions specified above, and is zero elsewhere. Parameter u was used as an auxiliary variable in order to obtain a left-skewed prior density for N. The rest of the model was specified by the equations
(7)
The observed weights of the samples were m*=451 360g, m1=4200g, m2=4300g and m3=4500g. Posterior distributions for,μ δand N were calculated by using Win BUGS. The posterior distribution for N describes the uncertainty about the number of fish in the rearing pond (Figure 1a). The 95% probability interval (PI) of the number of fish in the tank is [11130, 12030], and the most probable number (maximum a posteriori (MAP) estimate) is about 11570. If the group of fish in the tank is meant to be a mandatory release group of at least 12000 smolts, it might be of interest to calculate the probability that there are 12000 fish or more in the tank. This can be calculated during the MCMC simulation or from the resulting posterior density. In this case the probability is 0.03, carrying the message that it is unlikely that the targeted release number would have been reached. Nearly identical results were obtained by assuming that the individual weights are Normally distributed (PI=[11 090,12 020], MAP=11 550, P (N 12 000, data) = 0.03) (Figure 1). The posterior distribution for the mean weight μ is also very informative compared to its prior distribution (Figure 1b). However, the posterior distribution of the coefficient of variation δ has not been updated from its prior distribution as strongly as the other parameters (Figure 1c). This reflects the fact that only three subsamples were to be used for calibration, with the consequence that there is not much information about the variance of the weight of individual fish in this data set. It also emphasizes the importance of prior information in situations in which the data are sparse.
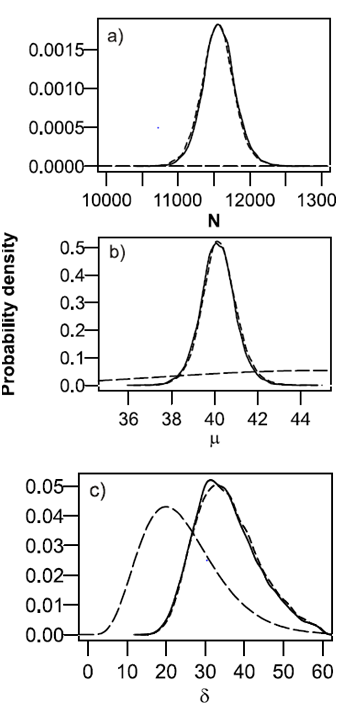
Figure 1 Posterior distributions, obtained by using a Gamma model (solid line) and a Normal model (short dashed line), and prior distributions (dashed line) of the number (N) of fish in the tank (a), of the mean
Posterior distributions, obtained by using a Gamma model (solid line) and a Normal model (short dashed line), and prior distributions (dashed line) of the number (N) of fish in the tank (a), of the mean ( μ) weight (b), and of the coefficient of variation (δ ) of the weight (c), based on the knowledge about the total weight of fish in the rearing pond and on information from three subsamples.
Discussion
Why Bayesian?
The model presented in this paper endeavors to answer a simple question: “Given my past experience and samples obtained, what should I think about the number of individuals in the large sample?”. Basing on the idea to use the concept of probability as a measure of personal degree of belief, the Bayesian approach is capable of answering such a question. All we need to do is to formalise in terms of probability what our past experience says about the distribution of the weight and about the number of individuals. The update of beliefs is obtained by applying the rules of probability calculus, and a quantitative answer to the original question is obtained in the form of the posterior distribution for the number of individuals in the large sample, providing an updated degree of belief in each possible value of the number of individuals. The frequentist approach, however, cannot provide a quantitative answer to the question. It is well known by statisticians, but not equally well appreciated by many applied scientists, that the frequentist approach deals only with the conditional distribution of observations given that the parameter values were known. The question for which the frequentist approach does provide a formal answer could then be stated as: “Given my past experience and a conjectured number of individuals in the large sample, what kind of samples could I expect to see if I repeatedly sampled the population for a very large number of times”. This question is quite different from the direct question concerning the unknown correct number of individuals in the sample. However, these two questions are obviously related, as it makes sense to believe more in numbers that would lead to data like those observed more frequently than in numbers that would make the observed data look more rare under the assumed sampling distribution. Thus, the result of the frequentist analysis can be intuitively connected to the question of actual interest, but the idea of direct probabilistic inference about the unknown number of individuals is lost.
Why not to compare to frequentist results?
The numerical values of the frequentist confidence intervals and point estimates may sometimes be close to those of corresponding Bayesian posterior probability intervals and MAP estimates. This does not mean, however, that the choice of the approach would then not matter8 despite the similar values they are answers to different questions. Existence of such claims indicates that the results of one approach or the other have been misinterpreted. More commonly, the results of a frequentist analysis are interprested as if they were the results of a Bayesian analysis.9 For the above reasons we argue that direct comparison between the results of Bayesian and frequentist analysis is pointless. However, many scientific journals seem to insist on such a comparison when results of Bayesian models are presented, thus increasing the risk that the conceptual differences between the two approaches become completely confused.
Why bother to specify informative priors?
It might seem that the prior distributions of model parameters did not have much influence on the resulting inference about the unknown size of the large sample, because prior distributions of the mean weight and the population size happened to be relatively flat compared to resulting posterior distribution. However, the prior for the coefficient of variation has been important for the resulting posterior. The marginal likelihoods of the mean weight and the population size both obviously depend on the information about the variation of the weight and the observed data did not contain much information about that. Being wise afterwards, one could claim that it would have been sufficient to elicit expert information only about the variation of the weight and only specify vague priors for the other parameters. In this case the conclusions about the population size would have been practically the same, but the key thing to note is that it really only applies to this particular data and initial information. In order to provide honest updating of knowledge, the prior distributions and the model structure should be specified to reflect the state of information before obtaining data. At that stage it is unknown what kind of data points will be observed, and thus it is unknown how much the posterior distribution will in the end depend on the prior opinion.
Vague or reference priors have often been suggested to make the Bayesian analysis objective and to let data to speak for themselves, or to represent initial lack of information. At least in the context of the problem dealt in this paper, such ideas would lead to quite obscure situations. In any conceivable real application of the model presented here, the researcher using the model will know what items she or he is considering. Thus, depending on the details (species and age of animals, for example) given about the items and on her or his past experience about the items, there will be some information about the mean weight and the variation of the weight, as well as about the shape of the weight distribution. The fact that statistical inference about the number of individuals is required already tells that it is thought to be so large that it is not worthwhile to try count the items exactly. Would the inference about the number of individuals become independent of the researcher’s beliefs (objective) if she or he used vague reference priors as if pretending to know nothing about the number individuals, their mean weight and the variation of the weight? Obviously not. The inference would then be dominated by the likelihood function, which is just a statement of her or his conditional prior beliefs about data given the parameter values and viewed as a function of parameters.10 The role of this subjective assumption about the shape of the weight distribution becomes more and more important as the number of samples taken from the population increases because the likelihoods imposed by each data point are multiplied with each other. Thus, there is no way around subjectivity in this context, nor in the statistical analysis as a whole.
Further development
The Bayesian model presented here can be used as a building block in more complex Bayesian models. For example, a model which describes the survival, harvest and reproduction of reared fish would need this type of model structure for the estimation of the number of stocked fish, the number of fish caught and the number of eggs from gonad samples. It could also be plugged into a stochastic VPA11, 12 to account for uncertainty about catches. When subsamples consist of only a single fish each, the inferences will generally be sensitive to the assumed shape of the weight distribution. If in doubt, one could consider extending the present model and apply non parametrically defined weight distributions.13 However, when each subsample contains larger amounts of fish, the assumed shape no longer plays a major role. This is because, when the number of fish in a subsample increases, and regardless of the shape of the weight distribution, the distribution of the sum of the weights resembles more and more a Normal distribution. On the other hand, for right-skewed weight distributions and small sample sizes the Gamma distribution can be regarded as safer choice. In our example the number of fish in each subsample was large enough to make the results robust to the choice between Gamma and Normal distributions. If all individuals were assumed to be of equal weight, and only measurement error was assumed to be present, then the problem could be seen as an estimation of a ratio parameter and methods proposed by Raftery & Schweder14 could be used. Subsamples of different sizes can be used at the same time in the analysis. For example, individual weights and weights of subsamples consisting of hundreds of fish can be utilized jointly. Finally, prior distributions of model parameters can be given a hierarchical structure in order to transfer information between exchange-able units, like fish farms, rearing ponds, or spawners.
Parameterization of the gamma distribution
The Gamma distribution is parameterized in this paper in terms of the mean and the standard deviation. The probability density function of a Gamma distributed variable x is
Acknowledgments
This paper has been partly funded by EU projects”Creation of multiannual management plans for commitment” (project nr SSP8-CT-2003-502289) and ”Oper-ational evaluation tools for fisheries management options” (project nr SSP8-CT-2003-502516). The study was also supported by the Graduate School of Computational Biology, Bioinformatics and Biometry (ComBi) and by the centre of Population Genetic Analyses supported by the Academy of Finland (project nr 53297).
Conflicts of interest
References
- Katia O. Ornamental fish trade. INFOFISH International. 2001;3:14–17.
- Saxena A. Health coloration of fish. International Symposium on Aquatic Animal Health: Program and Abstracts. University of California, School of Veterinary Medicine, Davis, CA, USA. 1994;pp.94.
- Torrissen OJ. Pigmentation of salmonids: Interaction of astaxanthin and canthaxanthin on pigment deposition in rainbow trout. Aquaculture. 1989;79(1–4):363–374.
- Withers PC. Comparative Animal Physiology. Brook Cole–Tomson Learning. Saunders College Publishing/harcourt Brace Jovanovich College, USA. 1992;pp.94.
- Goodwin TW. Carotenoids in fish. In: The biochemistry of fish. Biochemical Society Symposia, USA.
- Hata M, Hata M. Studies on astaxanthin formation is some freshwater fishes. Tohoku. Journal of Agricultural Research. 1973;24(4):192–196.
- Duncan PL, Lovell RT. Natural and synthetic carotenoids enhance pigmentation of ornamental fish. Highlights of agricultural research, Alabama Agricultural Experiment Station. 1993;40:8.
- Storebakken T, P Foss, K Schiedt, et al. Carotenoids in the diets for salmonids IV. Pigmentation of Atlantic salmon with astaxanthin, astaxanthin dipalmitate and canthaxanthin. Aquaculture. 1987;65(3–5):279–292.
- Chatzifotis S, Pavlidis M, Jimeno CD, et al. The effect of different carotenoid sources on skin coloration of cultured red porgy (Pagrus pagrus). Aquaculture Research. 2005;36:1517–1525.
- Dharmaraj S, Dhevendaran K. Application of microbial carotenoids as a source of colouration and growth of ornamental fish Xiphophorus helleri. World Journal of Fish and Marine Sciences. 2011;3(2):137–144.
- Ho ALFC, Zong S, Lin J. Skin color retention after dietary carotenoid deprivation and dominance mediated skin coloration in clown anemonefish, Amphiprion ocellaris. AACL Bioflux. 2014;7(2):103–115.
- Sinha A, OA Asimi. China rose (Hibiscus rosa sinensis) petals: a potent natural carotenoid source for goldfish (Carassius auratus L). Aquaculture Research. 2007;38(11):1123–1128.
- Theis A, Salzburger W, Egger B. The function of anal fin egg–spots in the cichlid fish Astatotilapia burtoni. PloS ONE. 2012;7(1):e29878.
- National Research Council (NRC). Nutrient requirements of fish. National Academy Press, Washington DC, USA. 1993.
- Czeczuga B, Dabrowski K, Rosch R, et al. Carotenoids in fish. Carotenoids in Coregonus lavaretus L. Individuals of various populations, Acta Ichth. Piscat. 1991;21(2):3–16.
- Foss P, Storebakken T, Liaaen Jensen S. Carotenoids in diets. V. Pigmentation of rainbow trout and sea trout with astaxanthin. Aquaculture. 1987;65(3–4):293–305.
- Ando S. Studies on the food biochemical aspects of changes in chum Salmon, Oncorhychus keta during spawning migration, mechanisms of muscle deterioration and nuptial coloration–Reprinted from memories of Faculty of Fisheries, Kokkaid University. 1986;33(1–2):1–95.
- Bjerkeng B, Storebakken T, Liaaen–Jensen S. Pigmentation of rainbow trout from start feeding to sexual maturation, Aquaculture. 1992;108(3–4):333–436.
- Wozniak M. Carotenoid contents in the body of rainbow trout Oncorhynchus mykiss, from different habitats. Fol Univ Agric Stetin 214 Piscaria. 2000;27:215–220.
- Castenmiller JJM, West CE. Bioavailability and bioconversion of carotenoids. Annu Rev Nutr. 1998;18:19–38.
- Furr HC, Clark RM. Intestinal absorption and tissue distribution of carotenoids. Nutritional Biochemistry. 1997;8(7):364–377.
- Tyssandier V, Lyan B, Borel P. Main factors governing the transfer of carotenoids from emulsion lipid droplets to micelles. Biochimica Biophysica Acta. 2001;1533(3):285–292.
- Tanaka Y. Comparative biochemical studies on carotenoids in aquatic animals. Mem Fac Fish. 1978;27(2):355–422.
- Torrissen OJ. Pigmentation of salmonids – a comparison of astaxanthin and canthaxanthin as pigment sources for rainbow trout. Aquaculture. 1986;53(3–4):271–278.
- Al–Khalifa AS, Simpson KL. Metabolism of astaxanthin in the rainbow trout (Salmo gairdneri). Comparative Biochemistry and Physiology. 198891(3):563–568.
- Torrissen OJ. Pigmentation of salmonids: Interaction of astaxanthin and canthaxanthin on pigment deposition in rainbow trout. Aquaculture. 1989;79(1–4):363–374.
- White DA, Page GI, Swaile J, et al. Effect of esterification on the absorption of astaxanthin in rainbow trout, Oncorhynchus mykiss (Walburn). Aquaculture Research. 2002;33:343–350.
- March BE, Hajen WE, Deacon G, et al. Intestinal absorption of astaxanthin, plasma astaxanthin concentration, body weight, and metabolic rate as determination of flesh pigmentation in salmonids fish. Aquaculture. 1990;90(3–4):313–322.
- Choubert G, Milicua JC, Gomez R. The transport of astaxanthin in immature rainbow trout Oncorhynchus mykiss serum. Comparative Biochemistry and Physiology. 1994;108(2–3):245–248.
- Parker RS. Absorption, metabolism and transport of carotenoids. FASEB J. 1996;10(5):542–551.
- Storebakken T, Hong KN. Pigmentation of rainbow trout. Aquaculture. 1992;100(1–3):209–229.
- Hardy RW, Torrissen OJ, Scott TM. Absorption and distribution of C–labelled canthaxanthin in rainbow trout (Oncorhynchus mykiss). Aquaculture. 1990;87(3–4):331–340.
- Aas GH, Bjerkeng B, Storebakken T, et al. Blood appearance, metabolic transformation and plasma transport proteins of C–astaxanthin in Atlantic salmon (Salmo salar L.). Fish Physiology and Biochemistry. 1999;21(4):325–334.
- Matsuno T, Tsushima M, Maoka T. Salmoxanthin, deepoxy–salmoxanthin and 7,8– didehydrodeepoxy–salmoxanthin from salmon Oncorhynchus keta. J Nat Prod. 2001;64(4):507–510.
- Gouveia L, Rema P, Pereira O, et al. Colouring ornamental fish (Cyprinus carpio and Carassius auratus) with micro algal. Aquaculture Nutrition. 2003;9(2):123–129.
- Ezhil J, Jeyanthi C, Narayanan M. Effect of formulated pigmented feed on colour changes and growth of red swordtail, Xiphophorus helleri. Turkish Journal of Fisheries and Aquatic Sciences. 2008;8(1):99–101.
- Schiedt K. Absorption and metabolism of carotenoids in birds, fish and crustaceans. In: Carotenoids Biosynthesis and Metabolism. Britton GS & Pfander H (Eds.), Birkhäuser: Basel, Switzerland. 1998;pp.285–358.
- Huyghebaert G. The utilisation of oxy–carotenoids for egg yolk pigmentation. Thesis of the Univiversity of Gent (Belgium). 1993.
- Seemann M. Eidotterpigmentierung: Unterschiede bei natürlichen und synthetischen Carotinoiden? DGS Magazin. 1997;49(36):24–28.
- Grashorn MA, Steinberg W, Blanch A. Effects of canthaxanthin and saponified capsanthin/capsorubin in layer diets on yolk pigmentation in fresh and boiled eggs. XXI World’s Poultry Congress, Canada. 2000;20–24.
- Andrewes AG, Starr MP. (3R, 3´R)–astaxanthin from the yeast Phaffia rhodozyma. Phytochemistry. 1976;15(6):1009–1011.
- Ako H, Tamaru CS, Asano L, et al. Achieving natural coloration in fish finder culture. In: Spawning and maturation of aquaculture species, Proceeding of the 28th UNJR aquaculture panel symposium, Kihei, Hawaii. 10–12, U NJR Tech Rep. 2000;28:1–4.

©2016 , et al. This is an open access article distributed under the terms of the,
which
permits unrestricted use, distribution, and build upon your work non-commercially.



