Journal of
eISSN: 2378-3184


Research Article Volume 2 Issue 3
1Department of Fisheries Biology and Genetics, Bangladesh Agricultural University, Bangladesh
2Department of Aquaculture, Bangladesh Agricultural University, Bangladesh
3Laboratory of Marine Biotechnology, University Putra Malaysia, Malaysia
4School of Fisheries and Aquacultural Sciences, University Malaysia Terengganu, Malaysia
Correspondence: Md Asaduzzaman, School of Fisheries and Aquaculture Sciences, University Malaysia Terengganu, 21030 Kuala-Terengganu, Terengganu, Malaysia, Tel +609-668-5041
Received: January 01, 2015 | Published: July 23, 2015
Citation: Shakur AAK, Haque MM, Rahman AM, Asaduzzaman M (2015) Morpho-genetic Analysis of Three River Populations of Bhagna, Labeo ariza (Hamilton 1807) in Bangladesh. J Aquac Mar Biol 2(3): 00029. DOI: 10.15406/jamb.2015.02.00029
Morphological and genetical analysis of Bhagna Labeoariza H. populations was investigated from three river populations (the Atrai, the Jamuna and the Brahmaputra) of Bangladesh to identify its diversified populations. Therefore, about 30 similar size of fish samples were collected from each of those three river populations and analyzed nine morphometric and meristic characters and their genetic variation by allozyme markers. All of the morphometric characters were significantly higher in (P<0.05) in Atrai population compare to other two river populations. However, among nine meristic characters, five were showed significant differences (P<0.05) among each other. The genetic variations were analyzed with five enzymes (EST, PGM, GPI, LDH and MDH) in CA 6.1 buffer system. Two polymorphic loci Est-1* and Pgm* were interpretable in muscle with starch gel electrophoresis. The highest mean number of allele per locus was observed in the Atrai population (1.375). The mean proportion of heterozygous loci per individual was 4.305% for all populations. The average observed heterozygosity (Ho) and expected heterozygosity (He) were 0.042 and 0.058, respectively for all the populations. The lowest pair-wise population differentiation (FST) (0.039) and the highest gene flow (Nm) (6.148) were found in the Jamuna–Brahmaputra indicating close relationship among them. In the Nei’s (1972) UPGMA dendrogram, the Jamuna and the Brahmaputra populations of L. ariza made one cluster (D=0.004) and separated from the Atrai population by the genetic distance of 0.066. The results suggest that a considerable genetic variation is existed among the natural L. ariza populations in Bangladesh.
Keywords: Labeoariza L, Morphology, Genetic variation, Dendrogram, River population
ANOVA, Analysis Of Variances; AFR, Anal Fin Rays; BR, Branchiostegal Rays; CFR, Caudal Fin Rays; DFR, Dorsal Fin Rays; ED, Eye Diameter; FL, Fork Length; GPI, Glucose-6-Phosphate Isomerase; HL, Head Length; HBD, Highest Body Depth; LBD, Low Body Depth, LDH, Lactate Dehydrogenase; MDH, Malate Dehydrogenase; Pr-OL, Pre-Orbital Length; PDL, Pre-Dorsal Length; PCFR, Pectoral Fin Rays; PEFR, Pelvic Fin Rays; PGM, Phosphoglucomutase; SaLL, Scale Along the Lateral Line; SL, Standard Length; SabLL, Scale Above the Lateral Line; SbLL, Scale Below the Lateral Line; SPSS, Statistical Social Package Program ; TL, Total Length
Labeoariza (Hamilton 1807) is commonly identified as Bhagna, Reba carp, Bhagna bata, Raik or Tatkini and locally it is regarded as one of the most desired indigenous minor carp species in Bangladesh. Bhagna is distributed over Indo-pacific region.1c-4 Bhagna additionally encompasses a wide range of geographical distribution. Primarily, this fish is found in Asia: Indus plain and adjoining hilly areas (Pakistan); Ganges-Brahmaputra basin (India, Nepal and Bangladesh); Mahanadi, Krishna and some more diminutive basins in southern India; Karnafuly and adjacent more diminutive basins in Chittagong Hill Tracts (Bangladesh).4-6 As well, Bhagna is widely distributed in most of the small creeks, rivers, floodplains and natural depressions of Bangladesh.7,8
Bhagna can be facilely apperceived by its consequential characteristics like the lower body is shiny and silvery; the upper body is deep bluish or darkish and medium-sized silvery scales. The lateral line consists of 38 or 39 scales and most of them appeared as elongated ebony marks from tail to the base margin of the operculum during the early stage of life. During the adult stage, the ebony mark becomes shorter and remains in between the terminus of the belly and the operculum.2,3,7-9 The thin stripes or a black mark is varying from subdued to bold. The dorsal profile is relatively more convex than the abdomen and, margin of snout is projecting, ciliated and covering upper lip completely and snout obtusely rounded. On the other hand, lower lip is very deficient and thin and is connected with skin of isthmus by a bridge and the upper lip is entire. The post labial groove is absent. The lower jaw is provided with a tubercle at symphysis. A pair of short rostral barbels present and mouth is inferior. Scales are hexagonal. Dorsal fin originates in advance of pelvics, midway between snout tip and posterior end of anal base. Pelvic fin origin nearer to snout tip than to caudal base. Pectoral fin shorter than head by less than eye-diameter. Longest dorsal ray may be slightly longer than head 3,9
Bhagna feeds on plankton and detritus and is an omnivore and column feeder in nature and grows to a largest size of 40 to 50 cm in length.8,10,11 This fish additionally grows up to 30.0 cm in standard length and weight of 1,360 g. Females and males procure sexual maturity within one year and customarily by the terminus of first year. This fish breeds in flooded shallow water from June to September.5The spawning season of Bhagna ranges from April to the August with a peak spawning time during the rainy time in flowing flood waters of these months.7,10 It is also a prolific breeder, laying about 3 million ova per female.5 Absolute fecundity of Bhagna was recorded between 2, 00,000 and 2, 50,000 per kg body weight of female .7
It is an eco-friendly fish and an important source of carbohydrates, proteins, fats, vitamins, minerals, iron and calcium.12 Its flesh is oily, tasteful and people would like to eat due to its lucrative size, high nutrition, attractive flavor and less spines.5,9 It is a congruous species for aquaculture.5,9 but the wild population of Bhagna is rapidly declining due to sundry causes. Over the last few years, the population of Bhagna has been declining considerably due to incremented and indiscriminant fishing pressure and sundry anthropogenic activities leading to aquatic pollution, siltation and loss of natural habitat for the magnification and spawning.7,9,10 These activities are perpetually eradicating the spawning and breeding grounds and causing havoc to the availability of fry, fingerlings and brood fish.7 As a result, the fish is recently listed as one of the most paramount vulnerably susceptible and threatened species in Bangladesh .13-15 Although they are listed as threatened and vulnerable, a small number of the fish is still available in different rivers, beels, haors and baors.15 For these reasons, it is necessary to conserve and rehabilitable through breeding and culture practices. Correspondingly, with the development of breeding and culture practices, information will be generated for promoting effective management and conservation practices and that will be helpful to protect the fish from extinction. So, immediate attempts should be taken to boost up Bhagna production through effective management in natural water bodies and a proper knowledge about their morpho-genetic variation is necessary.
During 1960s, molecular genetics studies on proteins such as hemoglobin and transferrin but attentions quickly turned to enzymatic proteins (allozyme/isozyme variation) on which most subsequent studies have been based.16 Since, fish geneticists have been utilizing protein electrophoresis as their primary implement to characterize population-level genetic variation in a variety of fish species.17 Isozymes the elements of different alleles at the same locus are named as allozymes. Allozyme electrophoresis represents the process for identifying genetic variation at the level of enzymes, which are directly encoded by DNA. The allelic variants give elevate to protein variants called allozymes that differ marginally in electric charges. Allozymes are co-dominant Mendelian characters (both alleles are individually expressed in a heterozygous individual) and characters are passed from parent to offspring in a prognosticable manner.18 Though co-dominant marker, they are functional for analyzing genetic variability and genetic identification of species.19,20 Fish population structure identification of several species has been carried out using allozyme technique.21-25 Allozymes were also found to be supportive in generating species-specific profiles and resolving taxonomic ambiguities in several species.26-28In this regard, Allozyme electrophoresis an effective tool for fish population studies and fishery management.29 However, there is no known information on stock identification and genetic variation of L. ariza in Bangladesh. Therefore, the present research was concentrated to investigate the morpho-genetic variation of the three major river populations of L. ariza regarding their present status to identify the diversified L. ariza populations through using allozyme markers.
Fish specimens
About 30 experimental fish were collected from each of three rivers viz. the Atrai (Dinajpur district), the
Brahamaputra (Mymensingh district) and the Jamuna (Sirajganj district) River in Bangladesh (Table 1).
|
Sample No. |
Populations |
District (Collection Site ) |
No. of specimens |
|
1 |
Atrai River |
Dinajpur (Chirirbandar) |
30 |
|
2 |
Brahmaputra River |
Mymensingh (Sadar) |
30 |
|
3 |
Jamuna River |
Sirajganj (Kajipur) |
30 |
Table 1 Sources and number of L. ariza samples collected from three river systems of Bangladesh
Morphological variation study
The conventional method was used to measure the nine morphometric characters as noted: Total Length (TL); Fork Length (FL); Standard Length (SL); Head Length (HL); Eye Diameter (ED); Pre-Orbital Length (Pr-OL); Pre-Dorsal Length (PDL); The Highest Body Depth (HBD); Low Body Depth (LBD) To The Nearest 0.1 Cm Using A Slide Calipers. Nine Meristic Characters Were Also Studied By Following The Conventional Methods: Branchiostegal Rays (BR); Dorsal Fin Rays (DFR); Pectoral Fin Rays (PCFR); Pelvic Fin Rays (PEFR); Anal Fin Rays (AFR), Caudal Fin Rays (CFR), Scale Above The Lateral Line (SabLL), Scale Below The Lateral Line (SbLL) And Scale Along The Lateral Line (SaLL). A magnifying glass was used to count the fin rays and only the principal rays were counted as separate ray.
Morphological data analyses
Due to limited number of fish in each group, non-parametric statistical analysis was used in all the comparisons for morphological data analysis .30 Kruskal-Wallis non-parametric analysis of variance (ANOVA) was used to analyze the differences in morphometric characters and meristic counts of fish .31 In this instance a non-parametric post hoc test.30 was conducted where significant differences between groups were detected. SPSS (version 12) program software was used to analyze all of the data.
Genetic variation study
About 30 fish were used to analyze the genetic variations for each river populations. Allozyme markers through horizontal starch gel electrophoresis were used to perform this study.32 The enzymes analyzed and enzyme commission (EC) numbers are shown in Table 2. Allozyme electrophoresis was performed by using amine-citrate buffers (CA 6.1).33 (Table 3). The components of staining solution with some co-factors that were used in this experiment are listed in Table 4. Gel slices (1 mm) were stained histochemically for different enzymes.34
|
Enzymes |
Enzyme Patterns |
E.C. Number |
|
Esterase (EST) |
Monomer |
3.1.1.1 |
|
Glucose-6-Phosphate Isomerase (GPI) |
Dimer |
5.3.1.9 |
|
Lactate Dehydrogenase (LDH) |
Tetramer |
1.1.1.27 |
|
Malate Dehydrogenase (MDH) |
Dimer |
1.1.1.37 |
|
Phosphoglucomutase (PGM) |
Monomer |
5.4.2.2 |
Table 2 Examined enzymes used for allozyme electrophoresis
|
Buffer system |
Gel buffer |
Running time |
Voltage |
References |
|
CA 6.1 |
1:20 dilution of electrode buffer |
Thick gel |
100 V |
(Clayton & Tretiak, 1995) |
Table 3 Composition of buffer system and allozyme electrophoretic process
|
Enzyme Name |
PMS |
NBT |
MTT |
Cofactor |
Other components |
|
EST |
- |
- |
- |
- |
0.1M Phosphate buffer*1 (60 ml) |
|
GPI |
* |
- |
* |
NADP |
0.05 M Tris-HCl*2 (80 ml) |
|
LDH |
* |
* |
- |
NAD |
0.05 M Tris-HCl*2 (80 ml) |
|
MDH |
* |
* |
- |
NAD |
0.05 M Tris-HCl*2 (80 ml) |
|
PGM |
* |
- |
* |
NADP |
0.05 M Tris-HCl*2(80 ml) |
Table 4 Components of staining buffer used for allozyme electrophoresis
*: After use showed activity; *1: Phosphate Buffer (Ph 7.0): Mixture of 0.2 M Sodium Dihydrogen Phosphate and 0.2 M Disodium Hydrogen Phosphate; *2Trishcl Buffer (pH 8.7): Tris (Hydroxymethyl) Aminomethane, pH Adjusted with 1N HCl; NAD: Β Nicotinamide Adenine Dinucleotide; NADP: Β-Nicotinamide Adenine, Dinucleotide Phosphate, Sodium Salt; NBT: Nitro Blue Tetrazolium; MTT: (3-[4, 5 Dimethyl-2-Thiazolyl]-2, 5 Diphenyl Tetrazolium Bromide); PMS: Phenazine MethoSulfate.
Genetic variation data analyses
The allele frequency and the analyses of Chi-square (χ2) test were performed using Gene Alex.35 computer program (version 6). The POPGENE (version 1.31) computer package program was used to determine the mean proportion of polymorphic loci.36 The mean number of allele per locus was determined and calculated directly from the number of observed alleles by dividing with the number of observed locus. The mean proportions of heterozygous loci per individual were calculated directly from total number of observed heterozygote loci by dividing with the total number of individual in a population. POPGENE (version 1.31) computer package program was also used to calculate the expected heterozygosity (He), observed heterozygosity (Ho), population differentiation (FST) and gene flow (Nm). Genetic distance values (D) was estimated through.37 Based on the D-values, dendrogram was made by the UPGMA (unweighted pair group method using arithmetic average) method.37
Morphological variation observations
The recorded nine morphometric characters from the samples of three river populations of L. ariza shown in the Table 5. The observed all nine morphometric characters of the Atrai population demonstrated significantly higher average values compared to the other two populations (the Jamuna& Brahmaputra). The proportion of five morphometric characters (TL: HL, SL: HL, ED: HL, SL: HBD and TL: HBD) of all river populations were significantly different (P<0.05) from each other. The recorded nine meristic characters from the three river populations of L. ariza are shown in Table 6. Of the nine meristic characters, four characters (BR, VFR, SabLL and SbLL) of all river populations showed no significant difference (P>0.05) from each other. The number of DFR, PFR and SaLL of the Atrai and the Jamuna populations were significantly higher (P<0.05) than the Brahmaputra population. On the other hand, the number of CFR of the Brahmaputra population was significantly different from other two populations (Table 6).
|
Morphometric characters |
Atrai population |
Jamuna population |
Brahmaputra population |
|
Total Length (TL) |
16.21±0.42a |
14.20±0.33b |
13.87±0.47c |
|
Fork Length (FL) |
14.00±0.39a |
11.99±0.25c |
12.16±0.26b |
|
Standard Length (SL) |
13.03±0.39a |
11.19±0.59c |
11.22±0.41b |
|
Head Length (HL) |
2.85±0.14a |
2.84±0.11b |
2.35±0.16c |
|
Eye Diameter (ED) |
0.76±0.07a |
0.64±0.06b |
0.64±0.06b |
|
Pre Orbital Length (Pr-OL) |
0.82±0.12a |
0.82±0.06a |
0.64±0.16b |
|
Pre- Dorsal Length (PDL) |
5.56±0.18a |
4.97±0.17b |
4.71±0.23c |
|
Highest Body Depth (HBD) |
3.53±0.17a |
3.36±0.13b |
3.11±0.07c |
|
Low Body Depth (LBD) |
1.79±0.08a |
1.42±0.06b |
1.31±0.08c |
|
TL : HL |
6.00±0.01a |
5.00±0.10c |
5.90±0.10b |
|
SL: HL |
4.57±0.03b |
3.94±0.01c |
4.77±0.02a |
|
TL:HBD |
4.82±0.03a |
4.02±0.01c |
4.45±0.04b |
|
SL: HBD |
3.87±0.03a |
3.16±0.02c |
3.60±0.01b |
|
EL:HL |
4.50±0.05a |
4.43±0.03b |
3.67±0.04c |
Table 5 Morphometric characters (average ±SE) as recorded from L. ariza sample of three river populations in Bangladesh (n=100)
|
Meristic characters |
Atrai population |
Jamuna population |
Brahmaputra population |
|
Branchiostegal Rays (BR) |
3.00±0.00a |
3.00±0.00a |
3.00±0.00a |
|
Dorsal Fin Ray (DFR) |
11.03±0.18a |
10.86±0.57b |
10.26±0.52c |
|
Pectoral Fin Rays (PFR) |
16.23±0.43b |
16.26±0.44a |
16.00±0.00c |
|
Ventral Fin Ray (VFR) |
9.00±0.00a |
9.00±0.00a |
9.00±0.00a |
|
Anal Fin Ray (AFR) |
7.03±0.18a |
6.23±0.43c |
6.76±0.43b |
|
Caudal Fin Ray (CFR) |
18.60±0.72b |
18.26±0.44c |
18.70±0.46a |
|
Scale Along Lateral Line (SaLL) |
37.53±0.81a |
35.63±1.27b |
35.53±1.35c |
|
Scale Above Lateral Line (SabLL) |
7.50±0.00a |
7.50±0.00a |
7.50±0.00a |
|
Scale Below Lateral Line (SbLL) |
6.50±0.00a |
6.50±0.00a |
6.50±0.00a |
Table 6 Meristic characters (average ±SE) as recorded from L. ariza sample of three river populations in Bangladesh (n=100)
Genetic variation observations
The enzymes were controlled and regulated by the genes at 8 presumptive loci as indicated by the electrophoretic patterns of muscle tissue. One of the 8 loci, the Est-1* formed two genotypes (*aa and *ab) by two alleles (*a and *b), whereas only one locus Pgm* formed four genotypes (*aa, *ab, *bb and *bc) by three alleles (*a,*b and *c) (Table 7). Six loci viz. Mdh-1*, Mdh-2*, Ldh-1*, Ldh-2*, Gpi-1* and Gpi-2* formed only homozygous genotype (*aa) with fixed allele *a. On the average 2 genotypes were produced by 1.6 alleles at the 5 loci are shown in (Table 7). Allele frequencies were measured from observed genotypes at eight loci in 90 samples of three river populations (Table 8). Among the eight loci, two polymorphic loci (Est-1* and Pgm*) was identified from the Atrai and the Jamuna populations. On the other hand, only one polymorphic locus (Est-1*) was identified from the Brahmaputra population. The chi-square (χ2) test was done in all the cases of polymorphic loci between observed and expected genotypes, based on Hardy-Weinberg equilibrium. The probability of two polymorphic loci is also showed in Table 8. The significant variations in allele frequency of Pgm* and Est-1* loci were observed in The Atrai and the Jamuna populations, whereas other population did not show any significant variation in allele frequencies (Table 8). The monomeric enzyme esterase (EST) presumably controlled by at least 2 loci, Est-1* and Est-2* and exhibited two banding patterns consisting of two heterodimers and one homodimer (Figure 1). Allele *a frequency ranged from 0.850 to 0.950 and was dominant in all three populations. Allele *b frequencies ranged from 0.050 to 0.183 and was present in all three populations (Table 8). The Est-2* was not readable due to complex banding pattern. The single Pgm* locus controls the monomeric enzyme phosphoglucomutase (PGM) (Figure 2). The Pgm* was monomorphic in the Brahmaputra population and polymorphic in the Atrai and the Jamuna populations with the allelic frequency of *a was ranged from 0.300 to 1.000. Allele *b was also present in the Atrai and the Jamuna populations with frequencies ranged from 0.183 to 0.683 but it was absent in the Brahmaputra population (Table 8) where the rare allele *c (0.016) is present in the Atrai population (Figure 2).
|
Locus |
Alleles |
Genotypes |
||
|
No. |
Type |
No. |
Type |
|
|
Est-1* |
2.0 |
*a, *b |
3.0 |
*aa, *ab, *bb |
|
Pgm* |
3.0 |
*a, *b, *c |
4.0 |
*aa, *ab, *bb, *bc |
|
Ldh-1* |
1.0 |
*a |
1.0 |
*aa |
|
Mdh-1* |
1.0 |
*a |
1.0 |
*aa |
|
Gpi-1* |
1.0 |
*a |
1.0 |
*aa |
|
Average |
1.6 |
|
2.0 |
|
Table 7 Meristic characters (average ±SE) as recorded from L. ariza sample of three river populations in Bangladesh (n=100).
|
Locus |
Allele Frequency |
|||
|
Allele |
Atrai |
Jamuna |
Brahmaputra |
|
|
Est-1* |
*a |
0.950 |
0.816 |
0.850 |
|
|
*b |
0.050 |
0.183 |
0.150 |
|
P |
|
0.815 NS |
0.009*** |
0.365 NS |
|
χ2 |
|
0.054* |
6.673* |
0.818* |
|
Gpi-1* |
*a |
1.000 |
1.000 |
1.000 |
|
Gpi-2* |
*a |
1.000 |
1.000 |
1.000 |
|
χ2 |
|
0 |
0 |
0 |
|
Ldh-1* |
*a |
1.000 |
1.000 |
1.000 |
|
Ldh-2* |
*a |
1.000 |
1.000 |
1.000 |
|
χ2 |
|
0 |
0 |
0 |
|
Mdh-1* |
*a |
1.000 |
1.000 |
1.000 |
|
Mdh-2* |
*a |
1.000 |
1.000 |
1.000 |
|
χ2 |
|
0 |
0 |
0 |
|
Pgm* |
*a |
0.300 |
0.816 |
1.000 |
|
|
*b |
0.683 |
0.183 |
- |
|
|
*c |
0.016 |
- |
- |
|
P |
|
0.001*** |
0.932 NS |
NS |
|
χ2 |
|
14.925** |
0.007 |
0 |
Table 8 P: Probability of Chi-Square Value; *** Significant Level: P<0.01; ** Significant Level: P<0.05;
*Significant Level: P<0.10; NS: Non-Significant.
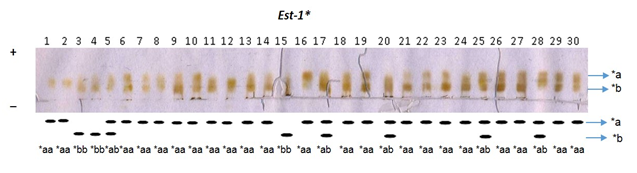
Figure 1 Electrophoregram of esterase (EST) and the schematic representation of electrophoretic patterns of Est-1* locus in the Jamuna population of Baghna.
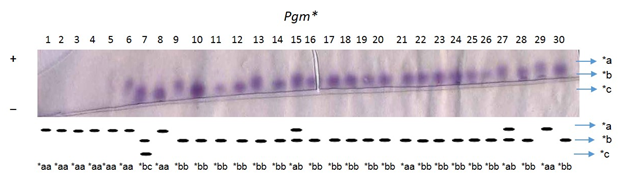
Figure 2 Electrophoregram of phosphoglucomutase (PGM) and the schematic representation of electrophoretic patterns of Pgm* locus in the Atrai population of Baghna.
The tetrameric enzyme lactate dehydrogenase (LDH) presumably controlled by two loci Ldh-1* and Ldh-2* and exhibited five banding pattern. The result indicated that the Ldh-1* and Ldh-2* loci were monomorphic and in all the three river population the allelic frequency of *a was found to be 1.00 (Table 8). The dimeric enzyme malate dehydrogenase (MDH) presumably controlled by at least two different loci, Mdh-1* and Mdh-2* and exhibited three banding patterns consisting of two heterodimers and one homodimer. The result also indicated that both the Mdh-1* and Mdh-2* were monomorphic and in all the three river population the allelic frequency of *a was found to be 1.00 (Table 8). The dimeric enzyme glucose-6-phosphate isomerase (GPI) presumably controlled by at least two different loci, Gpi-1* and Gpi-2* and exhibited three banding patterns consisting of one homodimer and two heterodimers, Both the Gpi-1* and Gpi-2* were also monomorphic and in all the three river population the allelic frequency of *a was found to be 1.00 (Table 8). In all the cases of polymorphic loci, the chi-square test based on Hardy-Weinberg equilibrium was carried out between observed and expected genotypes. Although the test was not effective in most of the cases in which the expected values were less than five, there were generally good agreements between observation and expectation (Table 8). However, significant variation in allele frequency of locus Est-1* was observe in the Atrai and Jamuna river populations. On the other hand, significant variation in allele frequency of locus Pgm* was observed in the Atrai population.
The genetic variability at 8 presumptive loci of L. ariza populations collected from 3 river population of Bangladesh is shown in Table 9. The mean fraction of polymorphic loci in the Atrai, Jamuna and Brahmaputra populations were 25, 25 and 12.5% respectively. The number of alleles per locus (Na) ranged from 1.125 (Brahmaputra) to 1.375 (Atrai) with a mean value of 1.250 for all populations. The mean fraction of heterozygous loci per individual was found to be 4.305% for all population. The observed heterozygosity (Ho) ranged from 0.033 (Atrai) to 0.058 (Jamuna) with a mean value of 0.042. The expected heterozygosity (He) ranged from 0.032 (Brahmaputra) to 0.076 (Jamuna) with a mean value of 0.058 for all population. The summary of the gene flow (Nm) and population differentiation (FST) are given in Table 10. The analysis of gene diversity within populations measured the gene flow (Nm) and the genetic differentiation (FST) over all the three river populations Bhagna are 0.652 and 0.277, respectively. In pair-wise analysis, relatively higher Nm value (6.148) was measured between the Jamuna and the Brahmaputra populations corresponding the lower level of FST value (0.039).
|
Population |
Mean Polymorphic Loci* (%) |
Mean Number of Alleles Per Locus (Na) |
The Mean Proportion of Heterozygous Loci Per Individual (%) |
Heterozygosity |
||
|
Ho |
He |
Ho/ He |
||||
|
Atrai |
25.000 |
1.375 |
3.333 |
0.033 |
0.068 |
0.485 |
|
Jamuna |
25.000 |
1.250 |
5.833 |
0.058 |
0.076 |
0.763 |
|
Brahmaputra |
12.500 |
1.125 |
3.750 |
0.037 |
0.032 |
1.156 |
|
Average |
20.830 |
1.250 |
4.305 |
0.042 |
0.058 |
0.801 |
Table 9 Genetic variabilities at 8 presumptive loci of L. ariza populations collected from 3 river population of Bangladesh
|
Populations |
FST |
Nm* |
||
|
Pair-Wise |
Overall |
Pair-Wise |
Overall |
|
|
Atrai-Jamuna |
0.207 |
0.277 |
0.956 |
0.652 |
|
Jamuna-Brahmaputra |
0.039 |
6.148 |
||
|
Atrai-Brahmaputra |
0.396 |
0.380 |
||
Table 10 Pair-wise and overall population differentiations (FST) and Gene flow (Nm) in three L. ariza populations.
* Nm = Gene flow estimated from FST = 0.25(1- FST) / FST
The genetic distance (D) values ranged from 0.004 to 0.037 among the three river populations (Table 11). The lowest genetic distance (D=0.004) was detected between the Brahmaputra and the Jamuna populations, while the highest value (D=0.066) was found in between the Atrai and the Brahmaputra populations. The UPGMA dendrogram constructed from Nei’s (1972) to show the genetic distances is shown in Figure 3. The UPGMA dendrogram displayed two clusters among the three river populations. The Jamuna and Brahmaputra population formed one cluster while the Atrai population formed another cluster. The dendrogram showed that the Atrai population separated from the Jamuna and the Brahmaputra populations by a genetic distance of 0.066 where the Jamuna population is different from the Brahmaputra population by the genetic distance of 0.004 (Table 11).
|
Populations |
Atrai |
Jamuna |
Brahmaputra |
|
Atrai |
*** |
0.962 |
0.935 |
|
Jamuna |
0.037 |
*** |
0.995 |
|
Brahmaputra |
0.066 |
0.004 |
*** |
Table 11 Nei's (1972) original measures of genetic identity (above diagonal) and genetic distance (below diagonal) estimated among 3 populations of L.ariza based on 8 loci
Morphological variation study
In this study, we collected all the samples at the similar time but higher total length of the Atrai population compared to that of the other populations might be due to relatively early natural breeding in that area due to natural climatic conditions. The offspring grow faster in early developing stage in natural habitat which might be the cause of growth variation in the Atrai population. In natural conditions, the growth of a particular species could be influenced by the different environmental factors which in turn, reflect the reproductive activity of that species.38 The proportion of total length and standard length of L. ariza with head length as obtained in the present study (5.00-6.00 and 3.00-4.77 times, respectively) is slightly lower than that reported in the previous study (5.8-6.3 and 4.5-4.8 times, respectively).3 In the present study, of the nine meristic characters of L. ariza of the Atrai, Jamuna and the Brahmaputra populations, the dorsal fin rays (11.03, 10.86 and 10.26), pectoral fin rays (16.23, 16.26 and 16.00), anal fin rays (7.03, 6.23 and 6.76), caudal fin rays (18.60, 18.26 and 18.70) and scale along lateral line (37.53, 35.63 and 35.53) were found to vary. These variations may be due to different environmental factors, geographical location of experimental population.
The present findings agreed with previous study by.39 who reported that the meristic counts of fishes are considered to be affected by environmental factors such as water temperature, pH etc in fresh water. The previous study reported 36-38 scales in lateral line of L. ariza populations and this meristic character strongly supported the present study.3 In the above mentioned observations, the morphological characteristic of all L. ariza populations seems to be somewhat similar. Although this species is already going to be threatened but their morphological characters are not destroyed. The observed morphological characters of L. ariza populations in Bangladesh showed minor variation from others and it is possibly due to the fact that their original sources might be from different locations. The mean values of each character diverse considerably by population, representing its exclusive characteristics .3
Genetic variation study
Five enzymes viz. GPI, LDH, EST, MDH and PGM were used in CA 6.1 buffer system and they produced clear resolution in the muscle tissue of the three populations of L. ariza. The enzyme GPI did not display clear resolution which might be due to the tissue and species specificity and utilized buffer system. If one has the information about the number of loci at which variation occurs (polymorphic loci), then only it would be possible to estimate the amount of genetic variation in a population.40 In order to monitor the levels of genetic variation in population, we can use the electrophoretic data because it provides such information of existence of polymorphic loci.41 The fraction of polymorphic loci (P) can be commonly used to estimate the electrophoretically noticeable variation in a population. Other commonly used measures of genetic variations are allele frequency in loci, mean number of allele per locus, average frequency of heterozygous loci per individual (H), observed heterozygosity (Ho) and expected heterozygosity (He).
In the present study, all populations showed two common alleles *a and *b except locus Pgm* in the Atrai population where a rare allele *c (0.033) was observed. Similarly a rare allele c* (0.05) in Pgm* was also reported by.42 in the natural population of rohu. The rare allele (*c=0.033) was also presented in the locus Est-1* of Puntiussarana population Shukair and Kangsha, Bangladesh.43 However, the presences of this rare allele in the Atrai population were the evidence of superiority of the Atrai population and absence of allele *c in other populations provided the evidence of the loss of genetic variability.
The average fraction of polymorphic loci per population of the Atrai and the Jamunariver were higher (25%) than the Brahmaputra (12.5) population which indicates the increase of gene pool diversity in the Baghna population of the Atrai and Jamuna River than the Brahmaputra population. It might be due to the geographical barrier between different stocks. The previous study estimated polymorphic loci (P) as 15.2% (P≤0.95) for polymorphism in fish in general.44 Therefore, the studied L. ariza population of the Atrai showed a decent level of polymorphism which close to the above mentioned range. The average heterozygous loci of 13.33% per individual within the range (6.67-13.33%) was obtained for the three populations of sharpunti (P. sarana).43 and for both hatchery and natural populations of rohu the value was reported to be lower than 15% .42 In our study, the mean number of heterozygous loci per individual (4.305%) indicated that the status of heterozygous loci remarkably remains in all the three river populations. The average observed heterozygosity (Ho) obtained in the present study (0.042) is lower than that (0.091).45 but similar to that (0.038-0.080) [46] in case of Clariasmacrocephalus. The higher observed and expected heterozygosity (Ho = 0.058 and He = 0.076) exhibited by the Jamuna population of Bhagna indicated that the gene pool was preserved effectually. The Ho values indicated that those corresponding to the Jamuna of L. ariza populations are closer to the average values of Ho = 0.055 obtained for teleosts. 47 It was stated that an average observed heterozygosity (Ho) value for teleost fishes was found to be 0.051.48 However, the He values (0.032-0.076) obtained in the present study do not exceed the range of values (He = 0.062 to 0.118) which are generally considered as the higher margins of genetic variability .44
The co-efficient of gene differentiation (FST) in all three L. ariza populations examined for all loci was 0.277. This result indicated the presence of population with a trivial genetic differentiation and the number of individuals that migrate from one population to another is high as revealed as Nm =0.652. Between the Jamuna-Brahmaputra populations, the pair-wise population gene flow was higher (6.148) than all other between population comparisons with corresponding the lowest FST value of 0.039. The FST value (0.277) of L. ariza populations as obtained in the present study is lower than that obtained for other freshwater fishes such as loach (0.774) .49 and freshwater Gobi (0.698).50 However, the present FST value indicates that there are little genetic differentiation exists among the populations.
The UPGMA dendrogram showed that the three populations can be congregated into two as revealed in the dendrogram (Figure 3). First group is consists of only a single Atrai population and separated by the D=0.066 from second one whereas second group consists of the Jamuna and Brahmaputra populations. In second group, it was observed that the Jamuna population was separated from the Brahmaputra population at D=0.004. It found that in a variety of animals, D is approximately 1.0 for inter species comparisons, around 0.1 for subspeciesand0.01 for local races. The D-value between subspecies is approximately 0.20.51 Considering from the above-mentioned criteria, the studied L. ariza may be categorized as local race or population.
Following above discussion of allozyme variabilities it is observed that the Atrai population showed higher genetic variation and followed by the Jamuna and the Brahmaputra populations. The higher genetic variation in the Atrai population is due to geographical distance because the Atrai River is geographically far from the Jamuna and the Brahmaputra rivers. On the other hand, the Jamuna River is geographically nearer to the Brahmaputra River. But in the present study, the Atrai sample was collected from the Chirrirbandar Upazilla (Dinajpur) where the there is less possibility of mixing with the Jamuna and the Brahmaputa populations.
The authors deeply acknowledge the contribution of the main supervisor Professor Dr. Md Mukhlesur Rahman Khan who passed away recently. The authors are also grateful for financial support to Ministry of NSICT (National Science and Information and Communication Technology) in Bangladesh for research fund to conduct the study and a small financial support from the Ministry of Higher Education (MOHE), Malaysia under the Fundamental Research Grant Scheme (FRGS).
None.

©2015 Shakur, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.