Journal of
eISSN: 2572-8466


Review Article Volume 8 Issue 3
1Instituto de Investigaciones Químico-Biológicas de la Universidad Michoacana de San Nicolás de Hidalgo, México
2Tecnológico de Monterrey, School of Engineering and Sciences, Campus Querétaro, México
Correspondence: Elda Beltrán Peña, Laboratorio de Transducción de Señales, Edificio B3, Instituto de Investigaciones Químico-Biológicas de la Universidad Michoacana de San Nicolás de Hidalgo. Ciudad Universitaria. Francisco J. Múgica s/n, C.P. 58030, Morelia, Michoacán, México
Received: April 16, 2021 | Published: May 7, 2021
Citation: Carrillo-Flores E, Pazos-Solis DM, Dìaz-Bellacetin FP, et al. TOR regulates plant development and plant-microorganism interactions. J Appl Biotechnol Bioeng. 2021;8(3):68-74. DOI: 10.15406/jabb.2021.08.00255
The adaptation of plants to their ever-changing environment denotes a remarkable plasticity of growth that generates organs throughout their life cycle, by the activation of a group of pluripotent cells known as shoot apical meristem and root apical meristem. The reactivation of cellular proliferation in both meristems by means of TOR, Target Of Rapamycin, depends on specific signals such as glucose and light. TOR showed a significant influence in plant growth, development and nutrient assimilation as well as in microorganism interactions such as infection resistance, plant differentiation and root node symbiosis. This review highlights the pathways and effects of TOR in the sensing of environmental signals throughout the maturing of different plant species.
Keywords: target of rapamycin, plant development, plant-microorganism interactions
ABA, abscisic Acid; AMPK, AMP-activated protein kinas; AtPDK1, phosphoinositide-dependent kinase 1; CRY1/2, cryptochromes 1 and 2; CYC-CDK, RBR cyclin; CDK, cyclin dependent kinase; DOI, Days of interaction; ETI, effector-triggered immunity; EZ, elongation zone; FHB, fusarium head blight; FKBP, rapamycin-binding; IT, infection thread; KIN10, arabidopsis kinase 10; MZ, meristematic zone; OC, organizing center; PGPR, plant-growth-promoting rhizobacteria; PhyB, phytochromes B; PLD, Phospholipase D; PYR1, pyrabactin resistance1; QC, quiescent center; RAM, root apical meristem; RAPTOR, regulatory-associated protein of mTOR; RBR, retino blastoma-related; SAM, shoot apical meristem; SCN, stem cell niche; SnRK1, sucrose-nonfermenting-related kinase1; TAP46, type 2A phosphatase associated protein; TOR, target of rapamycin
Target of Rapamycin, TOR, protein is a kinase evolutionarily conserved from yeasts to plants and mammals that has a central role in the sensing of nutritional and energetic state of cell among other environment signals. In mammals and yeasts, TOR acts as a catalytic component in two structurally and functionally distinct protein complexes called TORC and TORC2. The first, sensitive to rapamycin under favorable growth conditions promotes cell division, translation and ribosome biogenesis and represses catabolic processes such as autophagy.1 While the second one, insensitive to rapamycin, is involved in spatial cell growth regulation and in modulation of structure and polarity of the cytoskeleton.2,3 Although rapamycin was the first TOR inhibitor discovered, by decades it was reported that most plants were insensitive to this compound.4,5 Nonetheless, Xiong and Sheen,6 reported that under anaerobic growth conditions, Arabidopsis showed sensitivity to rapamycin. The adaptation of plants to their ever-changing environment denotes a remarkable growth plasticity that generates organs throughout their life cycle, by means of activation of SAM, Shoot Apical Meristem and RAM, Root Apical Meristem. In both meristems, cell division reactivation requires TOR participation, which depends on specific signals for each of them: SAM requires glucose and light, while RAM requires glucose.7 In addition to being the master modulator of growth, TOR has been described as a negative regulator of plant immunity by interfering with the activity of salicylic acid and jasmonic acid.8 On the other hand, Nanjareddy et al.,9 showed that TOR participates in Rhizobium interaction with bean roots. Moreover, it has been reported that decrease in TOR expression in Arabidopsis gives at plant resistance to fungus Fusarium graminearum.10 Recently, Sun et al.,11 observed that TOR signaling controls the infection in rice by the fungus M. oryzae. Although recently there have been many reviews of TOR role in plant development regulation and some reports of TOR effect during plant- microorganism interaction, in this article we make a brief presentation of the processes that regulate plant development, we discuss the recent researches on the involvement of TOR metabolic pathways in plant development regulation and its role during plant-microorganism interactions.
Plant development depends on proliferation, expansion and cell differentiation
Plant growth is modulated by nutrient availability through of regulation of three processes: proliferation, expansion and cell differentiation, which will be briefly described below. Post-embryonic growth is maintained through specialized organs known as SAM and RAM. The new organs originating from these meristems are located in the opposite poles of the plant (Figure 1a). During embryogenesis, SCN, Stem Cell Niche, is located in the centers of both meristems. The SCN of SAM is made up of an OC, organizing center, (quiescent cells that rarely divide) and the initial cells, which have the capacity to renovate and produce new organs and tissues indefinitely, and whose progeny moves laterally towards the peripheral zone, where they undergo rapid cell division and finally it differentiated, forming the lateral organs, vascular tissues and stem (Figure 1b).12,13 The RAM that is located in root tip, also presents an SCN, formed by the QC, Quiescent Center, surrounded by initial cells, which divide asymmetrically to generate different cellular layers of root, arranged concentrically around of longitudinal axis (Figure 1c).14 During root development, a balance is reached between division rate and cell differentiation rate. The MZ, Meristematic Zone, originates cells by division from primary root, which proceed towards EZ, Elongation Zone (Figure 1c), where their size increases up to twenty-fold due to rapid vacuolar expansion. Finally, the cells undergo a differentiation in maturation zone of radicular hairs, characterized by the elongation of some cells from the epidermis.15
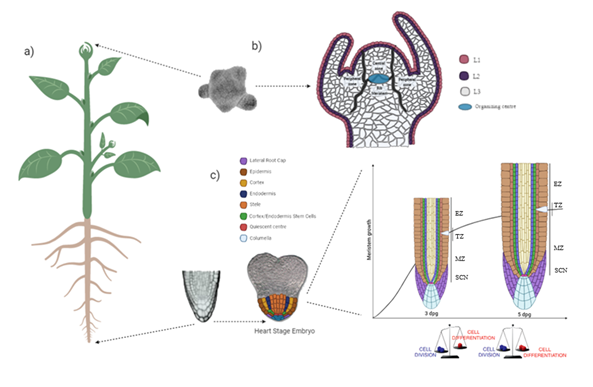
Figure 1 Structure and organization of SAM and RAM. a) Location of meristems in an adult plant. b) The diagram shows the different functional zones of SAM. c) RAM organization in heart stage embryo of Arabidopsis. Three dag, days after germination, root meristem size increases rapidly due to prevalence of division over cell differentiation, while at five dag this growth is achieved by an increase in differentiation rate that equals that of cell division.15,16 SCN, Stem Cell Niche; MZ, Meristematic Zone; TZ, Transition Zone and EZ, Elongation Zone.
Three dag, days after germination, root meristem size increases rapidly due to prevalence of division over cell differentiation, while at five dag this growth is achieved by an increase in differentiation rate that equals that of cell division.15,16 SCN, Stem Cell Niche; MZ, Meristematic Zone; TZ, Transition Zone and EZ, Elongation Zone.
TOR signaling pathway regulates growth and development plant
In plants, it was founded homologous proteins of some components of PI3K/TOR/S6K signaling pathway of mammals: P13K, Phosphatidyl Inositol 3-Kinase/TOR, Target Of Rapamycin/S6K, protein S6 Kinase (Figure 2a).17 In Arabidopsis thaliana, the homologous protein of PI3K is codified by the AtVPS34 gene, Vacuolar Protein Sorting 34, and AtVPS34 is activated by growth factors and is essential for growth.18,19 TOR is also activated by phosphatidic acid, which originates from of breakdown of a phospholipid membrane, by PLD, phospholipase D.20 Downstream of AtVPS34 we find AtPDK1 kinase, Phosphoinositide-Dependent Kinase 1, which regulates growth and proliferation cell21 and AtS6K phosphorylation.22 Another component is SnRK1, Sucrose-nonfermenting-Related Kinase1, a protein kinase that senses glucose/energy levels and is a plant ortholog of AMPK, AMP-activated Protein Kinase, in mammals. The signaling TOR-SnRK1 has a central and antagonistic function that impacts plant growth from early stages of development. SnRK1 is made up of one catalytic subunit (α) and two regulatory subunits (β, γ). KIN10, also known as Arabidopsis kinase 10 [AKIN10], one of the three isoforms of the catalytic subunit, is responsible for most of the SnRK1 activity; under conditions of energy deprivation and stress this protein interacts and phosphorylates at RAPTOR protein, Regulatory-Associated Protein of mTOR. In Arabidopsis, the overexpression of KIN10 results in late flowering, while the mutant causes early flowering.23 The SnRK1 complex can activate transcription factors bZIP2, bZIP11 and bZIP63 in response to starvation, leading to a stimulation of genes expression involved in catabolic pathways such as autophagy and the degradation pathways of cell wall components, starch, amino acids, sucrose, lipids and proteins; thereby providing alternative sources of energy and metabolites. The aforementioned suggests that SnRK1 regulates global plant metabolism, growth, and energy balance.24 The most studied target of TOR is S6K kinase; in Arabidopsis two genes, S6K1 and S6K2, were reported that encode proteins with an identity of 87%. It has been suggested that S6K2, is the protein homologous to S6K1 in mammals, since both phosphorylate ribosomal protein S6, thereby allowing increased selective recruitment of ribosomal protein transcripts to polysomes, thus affecting translational regulation.22 Furthermore, S6K2 activity in plants has been shown to increase in response to auxins and cytokinins.25 Recently, it was described that S6K, phosphorylated at RBR1 protein, Retino Blastoma-Related1, which stimulates its nuclear localization and inhibits of E2F transcription factor activity, and thus represses the transcription of genes that promote cell cycle (Figure 2a). Interestingly, Xiong et al.,26 reported that AtTOR kinase directly phosphorylates E2Fa, activating the transcription of genes of S phase of cell cycle; this discovery it deviates from the conventional CYC-CDK-RBR cascade, CYClin-Cyclin Dependent Kinase-Retino Blastoma Related, where the CYC-CDK complex phosphorylates at RBR and therefore releases transcription factor E2F. The aforementioned differs from the conventional function of TOR, where the stimulation of translation is involved in the cell cycle progression achieved through of S6K activation. Another TOR target is TAP46, Type 2A Phosphatase Associated Protein of 46 kDa, a regulatory subunit of protein phosphatase 2A that stimulates phosphorylation of S6K1. Plants that overexpress TAP46 show an increase in length of hypocotyl, of leaves and an increase in seeds size, as well as a high expression of genes associated with ribosome biogenesis, lignin biosynthesis and N assimilation. In contrast, translation and expression of genes involved in N assimilation are reduced and autophagy is induced in Tap46 RNAi lines.27
TOR is a highly conserved 250-kDa atypical serine/threonine protein kinase that belongs at PI3K family and is made up of the following domains: HEAT, Huntington repeats, Elongation factor 3 regulatory, subunit A of PP2A, TOR1; FAT, FRAM-ATM-TRAP; FRB, FKBP-Rapamycin-Binding, the catalytic kinase domain and the C-terminal FATC domain (Figure 2b). In various plant species, TOR is highly similar, especially in FRB and kinase domains. TOR senses energy and nutrient levels, and is a regulator of cell growth, in addition to modulating protein synthesis and metabolism. It is also involved in restoring cell wall integrity, proliferation, and cell size.28
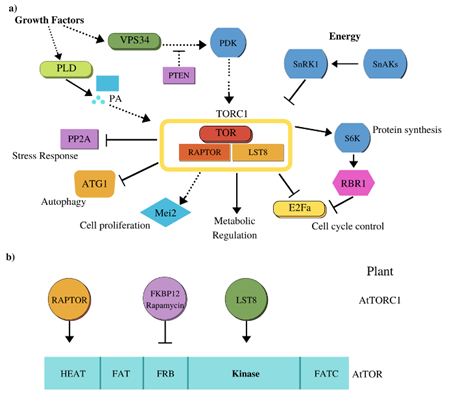
Figure 2 TOR signaling pathway in plants. a) AtVPS34 is activated by growth factors and by an unknown mechanism that phosphorylates PDK, to finally activate TOR, which then regulates protein synthesis, metabolism, cell proliferation, inhibits autophagy, stress response and controls cell cycle. b) Structure of TOR domains, highlighting the components that join each of them.17
Arabidopsis genome has a single gene for TOR and two for RAPTOR, whose products are components of the TORC1 complex. The study of TOR role in the development of plants was delayed compared to other organisms, since the mutation of this gene was lethal in early stages of A. thaliana embryogenesis. Deprost et al.,29 observed that growth, seed yield, resistance to osmotic stress, to ABA, abscisic acid, and sugar sensitivity correlated with AtTOR transcript levels (Figure 3a). Liu & Bassham30 observed in TOR mutants a constitutive autophagy and a decrease in protein synthesis, while that TOR overexpression caused an accelerated senescence.31 Menand et al.,4 using the TOR::GUS reporter line from Arabidopsis, showed that the protein is highly expressed in fast growing tissues such as SAM, embryo and endosperm,but not in differentiated cells. For several decades, plants have been reported to be insensitive to rapamycin, this was attributed to a variation in amino acid sequence of FBBP12 protein, which together with rapamycin binds to the FRB domain of TOR, thus inhibiting their activity. However, Xiong & Sheen6 reported for the first time that germinated of seeds and grown seedlings of A. thaliana under anaerobic conditions (liquid culture medium), showed sensitivity to rapamycin. Currently, the competitive inhibitors of ATP binding site of TOR kinase, known as second-generation inhibitors, are used in mammals. These competitive inhibitors can include: TORIN1, TORIN2, PP242, AZD-8055, KU-63794, WYE-132, WYE-354, QL-IX-55 e INK-128. To date, the following inhibitors have been tested in plants: AZD-8055, TORIN1, TORIN2, WYE-132, WYE-354 and KU-63794, with high effectiveness in blocking TOR with the consequent alteration of growth (Figure 3b).4,6,32–34
It has been reported that root hairs development is mediated by expression of LRX1, LRR-EXTENSIN1, and ROL5, REPRESSOR OF LRX, and that mutations of these latter gene in Arabidopsis are hypersensitive to rapamycin and show defects in cell wall, suggesting that TOR participates in cell wall integrity.35 Deng et al.,33 showed that plants treated with rapamycin and KU-63794, presented a decrease of auxin in RAM, in addition a significant decrease in TAA1, TRYPTOPHAN AMINOTRANSFERASE OF ARABIDOPSIS 1; TAR2, TAA2-RELATED PROTEINS; YUCCA1 and YUCCA2 genes expression involved in auxin biosynthesis, suggesting that TOR affects the auxin response pathway. Pfeiffer et al.,36 observed that in SAM, sucrose and red light promote the expression of WUS, which codes for a transcription factor that regulates at initial cells of OC. Genetic analysis revealed that participation of phyB, phytochromes B and CRY1/2, cryptochromes 1 and 2 in WUS activation by glucose or sucrose through energy signaling, is repressed by AZD-8055, suggesting that TOR could be involved in integration of energy and light signaling to promote the activation of initial SAM cells. On the other hand, Li et al.,7 reported that SAM proliferation reactivation depends on glucose and light, while that RAM only depends on glucose, and that TOR is involved in reactivation of both meristems (Figure 4a). Chen et al.,37 determined that light-activated phosphorylation of S6 stimulates protein synthesis and cotyledons opening and that TOR participates in both events (Figure 4(b&c).
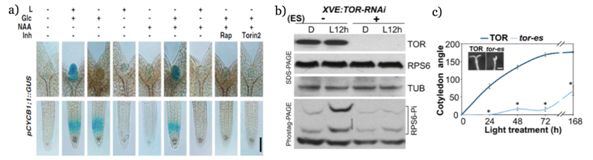
Figure 4 Reactivation of cell division of SAM and RAM and light-stimulated RPS6 phosphorylation depends on TOR. a) Glucose and light reactivate cell proliferation of both meristems, furthermore auxin replace at light in cell division reactivating along with glucose.7 b) 4-day-old seedlings of estradiol-conditioned line XVE:TOR-RNAi (Os), were grown with (+) and without (-) 20 µM of estradiol for 12 hours under direct light. Without TOR induction, light cannot induce RPS6 phosphorylation. c) Four-day-old etiolated seedlings of tor-es, estradiol inducible mutant grown with and without estradiol, were treated with 15 µE of white light to determine opening cotyledon for seven days. The tor-e seedling showed a significant delay in opening of these organs.37
Wu et al.,38 indicate that more and more plant-specific factors that induce TOR activation are being discovered, as well as new targets for this kinase. Some of them are exclusive to plants such as E2Fa phosphorylation, which promotes cell division, and PYR1/PYL/RCAR (Pyrabactin Resistance1(PYR1)/PYR1-Like/Regulatory Components of the ABA Receptor) family of proteins (herein referred to as PYL). PYL activation regulates seed germination and greening (Figure 5a). By phosphorylating of S6K1/2, TOR regulates protein translation involved in cotyledon opening and greening, among other events, while E2Fa/b phosphorylation stimulates meristem activation, leaf development and endo-reduplication (Figure 5b).38
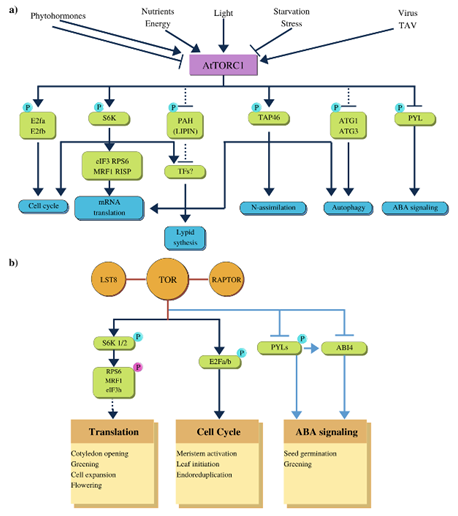
Figure 5 Plant TOR is activated by different effectors. a) TOR is activated by nutrients, energy, auxin and light, and inactivated by phytohormones and stress by inanition. TOR phosphorylates directly at E2Fa and E2Fb factors that regulate cell cycle. Another TOR target is S6K1, which upon activation by auxin phosphorylates at eIF3h, subunit of initiation factor eIF3, allowing translational reset of uORF-mRNAs. MRFI, MA3 DOMAIN-CONTAINING TRANSLATION REGULATORY FACTOR1, regulates translation of mRNAs in energy deficiency conditions.38 b) TORC1 complex is made up of LST8, TOR and RAPTOR proteins; TOR active phosphorylates at S6K1/2, E2Fa /b and PYL, involved in regulation of translation, cell cycle and ABA signaling respectively.38
It has been reported that photosynthesis or exogenous glucose supply stimulate signaling of TOR-S6K pathway.26 However, even in presence of glucose or sucrose, TOR pathway is not activated when nitrogen or phosphorus is missing, suggesting that these elements and/or their metabolic derivatives are signals for activation of such pathway. Even more, it appears that nitrate, ammonia, and amino-acids also participate in TOR-S6K pathway activation.38
Effect of the TOR pathway on the plant-microorganism interaction
Plant-bacteria association has been studied for several decades and it has been observed that the concentration of bacteria around the roots is generally higher than that found in the rest of soil. This has been attributed to the fact that exudates (representing 5 to 30% of photosynthetically fixed carbon) secreted by roots, are used as a carbon source by the bacteria. The mechanisms employed by soil bacteria to facilitate growth are well known in PGPR, Plant-Growth-Promoting Rhizobacteria.39–41 Due to their ability to colonize root surfaces and internal tissues of plant, PGPR can stimulate the plant growth directly by facilitating iron acquisition, nitrogen and phosphate from environment and by auxin, cytokines and ethylene synthesis or indirectly by limiting or preventing damage to plants by pathogens, including bacteria, fungi, and nematodes.41
On the other hand, legumes establish symbiotic interactions with nitrogen-fixing bacteria in soil, through specialized root structures known as nodules. The symbiotic program begins with a chemical dialogue between rhizobia and plants that lead to rhizobial production of node factors, inducing a series of responses in root hairs (oscillations in calcium concentration, symbiotic genes induction and curly hair to catch bacteria).42,43 Subsequently, the hydrolysis of cell wall and invagination of plasma membrane allows the (IT)infection threads formation, which cross hair cell base.9 Recently, it was reported that during the symbiotic interaction between Phaseolus vulgaris-Rhizobium tropici, TOR expression was observed throughout the development of IT and in infected cells of mature nodules; while TOR silencing, affected IT progression and division of associated cortical cells, resulting in a number of nodules drastic reduction. All these data suggest that TOR has a vital role in root node symbiosis establishing in beans.9 On the other hand, to survive the attack of microbial pathogens and herbivorous insects, plants have developed a response known as ETI, Effector-Triggered Immunity that prevents the colonization of the pathogen and represses the development of disease. The success of pathogen colonization is due to the fact that it interrupts ETI by injecting small effector proteins into of apoplast or host cytosol. The plants, in turn, have adapted to recognize the attacker's specific effectors through intracellular or transmembrane resistance proteins, which trigger the ETI.44,45 De Vleesschauwer et al.,8 used rice plants as a study model to investigate whether TOR modulates the pathogen-rice interaction, and observed in plants overexpressing TOR and RAPTOR (OsRaptor OX and OsTOR OX), long shoots, an increase root growth and epidermal and mesophilic cells size, an improvement in maturation, early flowering and higher seed yield, while Raptor RNAi lines showed a reduced root growth (Figure 6a). Subsequently, inoculation of rice plants of OsRaptor OX lines with the pathogenic bacteria Xanthomonas oryzae pv resulted, at 12 doi, days of interaction, a significantly higher susceptibility at hemibiotrophic bacteria Xoo, which causes leaf blight disease. In contrast, Raptor RNAi plants exhibited a 50 % reduction in lesion length. These results suggested that TOR acts as a negative regulator of resistance to Xoo. Fusarium, one of the most important genera of pathogenic fungi that causes the devastating disease known as Fusarium Head Blight (FHB) in wheat, barley and other cereals, is known to cause a decrease in grain quality and yield, in numerous crops of economic importance. Recently, Aznar et al.,10 evaluated the Arabidopsis-Fusarium graminearum interaction in raptor3g, lst8.1, lst8.2 mutant lines and in TOR mutant conditioned to estradiol tor-es. The rosette leaves of such plants were inoculated with a virulent Fusarium isolate, which caused symptoms of disease in wild plants. However, a resistance reaction was observed in the raptor3g and tor-es lines (Figure 6b). The results described above demonstrated that TOR signaling down-regulation confers plant resistance/tolerance to the Fusarium infection.
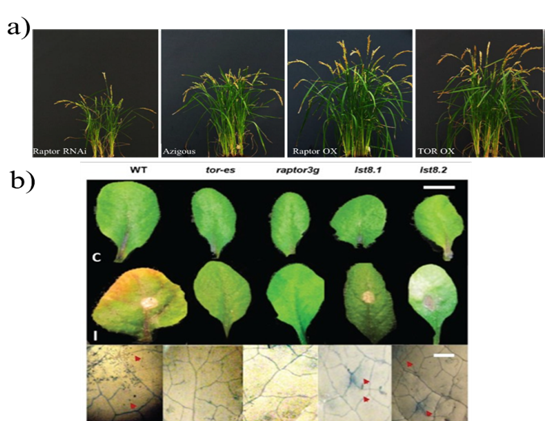
Figure 6 TOR regulates rice development and TORC1 complex mutants of Arabidopsis show resistance to Fusarium graminearum infection. a) Phenotypes of TOR overexpression rice lines. The photographs were taken 12 weeks after germination.8 b) Symptoms of Fusarium head blight disease in mutant lines of Arabidopsis TORC1 complex, is shown the evidence of lesions in Wt leaves and mutants at 7 doi with Fusarium compared to the control (I).10
From the aforementioned, it is clear that TOR activated by light, phytohormones, nutrients and energy regulates plant development through the control of cell cycle, transcripts translation involved in development and function of cotyledons, N and S assimilation, cell wall integrity, secondary metabolism and pathway ABA activation. In this review, we also describe how TOR participates in the adaptation of plants to infection with some fungi and bacteria, for example, down-regulation of TOR conferring at Arabidopsis a resistance to Fusarium infection, while in rice seedlings with mutations in some of components of complex TORC1, lesions size caused by infection with pathogens Xanthomonas oryzae and Magnaporthe oryzae was found to be reduced. Recently, two studies identified new TOR targets, in the first one, resistant mutants at inhibitors of TOR were isolated,46 and in the second a phosphoproteomic analysis identified new TOR targets.47
This work was supported by the Coordinación de la Investigación Científica UMSNH. ECF was fellow of CONACYT-México (7080155).
The author declares that there is no conflict of interest.
None.

©2021 Carrillo-Flores, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.