Journal of
eISSN: 2572-8466


Research Article Volume 9 Issue 5
1National Facility for Biopharmaceuticals, GN Khalsa College, India
2Department of Biotechnology, B.K. Birla College of Arts, Science & Commerce, India
3Department of Biotechnology, The Institute of Science, India
4Department of Biotechnology, Kishinchand Chellaram College, India
5Department of Biological Sciences, SVKM’s NMIMS Sunadan Divatia School of Science, India
Correspondence: Vikas Jha, National Facility for Biopharmaceuticals, G. N. Khalsa College, Mumbai, Maharashtra, India
Received: September 09, 2022 | Published: October 11, 2022
Citation: Jha V, Sahu A, Bhosale A, et al. Isolation, characterization, and identification of bioactive metabolite from soil derived Streptomyces polyrhachis AS07. J Appl Biotechnol Bioeng. 2022;9(5):158-167. DOI: 10.15406/jabb.2022.09.00305
The increasing stress caused by antibiotic resistance necessitates the need to explore novel and effective antimicrobial agents from actinomycetes which are often acknowledged for their capability to produce a variety of antibiotics. This study focuses on investigating the antimicrobial properties of the secondary metabolite synthesized by the soil resident Streptomyces polyrhachis AS07. The soil isolated strain was identified using 16S rRNA sequencing technology and phylogenetic analysis. Under appropriate laboratory conditions, the Streptomyces sp. produced sufficient yield of secondary metabolite for assessment of antimicrobial properties. The Gas Chromatography Mass Spectrometry chromatogram of the metabolite revealed abundant presence of antimicrobial constituents such as propionic acid, palmitic acid, and other compounds from tetrazolic and monocarboxylic groups. When tested against a broad range of Gram-positive and Gram-negative bacteria, the extracted metabolite exhibited appreciable antimicrobial activity with highest activity against S. aureus. Additionally, it also had potential antioxidant activity with an IC50 value of 5.50 μg/mL. The metabolite displayed highest antibiofilm activity against B. subtilis as well as successfully restricted the quorum sensing ability of C. violaceum. Furthermore, the extract significantly inhibited the swarming ability of P. aeruginosa upto 16.67 %. Conclusively, Streptomyces metabolite can be considered as a potent reservoir of bioactive molecules for industrial production with promising pharmaceutical applications.
Keywords: actinomycetes, antibiofilm, antimicrobial, antioxidant, GCMS, Streptomyces
WHO, World health organization; MRSA, methicillin-resistant staphylococcus aureus; GC, guanine cytosine; ISP, international streptomyces project; MK, menaquinone; DPPH, 2,2′-diphenyl-1-picrylhydrazyl; QS, quorum sensing; NFB, national facility for biopharmaceuticals; GCMS, gas chromatography mass spectrometry; EI, Electron Ionization; NIST, national institute of standard and technology; ATCC, American type culture collection; MTCC, Microbial Type Culture collection and gene bank; LB, luria-bertani; SDB, sabouraud dextrose broth; NCCS, national centre for cell science; LA, luria-bertani agar; SDA, sabouraud dextrose Agar; MIC, minimum inhibitory concentration; REMA, resazurin microtiter assay; UV, ultraviolet; OD, optical density; CFU, colony forming unit; ELISA, Enzyme-linked immunoassay; ANOVA, analysis of variance; IC, inhibitory concentration; ASB, ascorbic acid; ME, metabolic extract; DNA, deoxyribonucleic acid; MDRO, multi-drug resistant organism; PDR, pan-drug resistant; PDRO, pan-drug resistant Organism; MTSA, modified tryptic soy agar; NCBI, National Center for biotechnology information; PCR, polymerase chain reaction
Drug resistant infections have become a major health concern accounting for a global mortality rate of 17 million per year.1 According to 2019 WHO (World Health Organization) statistics, new anti-microbial resistances have been reported. One of the most notorious multi-drug resistant organism (MDRO) encountered till date is Methicillin-resistant Staphylococcus aureus (MRSA) which is known for its resistance not only against Methicillin but also against Aminoglycosides, Macrolides, Tetracycline, Chloramphenicol, Lincosamides as well as disinfecting chemicals.2 Secondly, the infamous pan-drug resistant (PDR) Pseudomonas aeruginosa has led to the ever-increasing global trendline of ocular infections.3 The resistance ability of this PDRO can be attributed to its capability to produce biofilm, an extracellular polysaccharide matrix, that acts as its defensive arsenal in hostile conditions.4 Another infectious PDR and gram-negative organism namely Acinetobacter baumanii has become well-known for higher mortality rates due to the absence of monotherapy treatment possibilities.5 In-spite ofhe discovery of new antimicrobials, antibiotic resistance is increasing at an alarming rate. Thus, the need for novel therapeutic drugs is in exclusive demand which will consist of enhanced modes of action that play a critical role in targeting the multidrug and pan-drug resistant microbial pathogens that are the principal cause of life-threatening infections.1
Streptomyces the largest genus of the Actinobacteria are one of the free-living residents found in soil and marine habitats that produce secondary metabolites consisting of numerous potential bioactive properties such as antibiotic, antifungal and antiviral activity and against a wide range of microorganisms.6 The organisms belonging to the genus Streptomyces are gram positive, spore forming, filamentous, soil inhabiting aerobes with high GC content and are the major microbial population present in soil.1 Streptomyces are considered to be the most versatile soil microbes and consist of biotransformation activities along with an ability to degrade lignocellulose and chitin as well as play a pivotal role in organic matter biological cycles.7 The organism belonging to this genus alone produces approximately three quarters of all known naturally occurring antibiotics worldwide.8 They have been shown to synthesize over 10,000 bioactive natural antibiotics, including Penicillin, Tetracycline, Gentamicin, Vancomycin And Pimaricin.9 A new study conducted by Djebbah et al.,10 performed an in-vitro antimicrobial analysis of the metabolic extract produced by the novel Streptomyces GLD22 strain. The extracted metabolite was reported to contain various bioactive molecules with antimicrobial, anti-inflammatory, antioxidant and cytotoxic activity. Another recent study by Qureshi et al.,11 has confirmed the antimicrobial properties of two actinomycin extracts X2 and D, both synthesized by the novel Streptomyces smyrnaeus UKAQ_23 strain isolated from the mangrove-sediment located in Saudi Arabia. Additionally, Quinn et al.,12 successfully isolated the novel Streptomyces CJ13 strain from soil used in Irish traditional medicine which was found to inhibit a broad range of multi resistant bacteria, anaerobes and fungi.
Although many studies have investigated the biological activities of the metabolic extracts synthesized by many other Streptomyces sp., only one study so far has reported the isolation of Streptomyces polyrhachis from the Chinese black ant (Polyrhachis vicina Roger).13 The obtained Streptomyces sp., an Actinomyces strain isolated from the Chinese black ant Polyrhachis dives, is rich in nutrients and has such numerous healthcare functions as the regulation of the immune system, relaxation of fatigue and anti-aging.13 They are able to grow on ISP3, ISP4, ISP6, ISP7, Czapek’s and nutrient agar and diffusible pigment melanin is observed on ISP3 and ISP4.14 They also contain menaquinones such as MK-8(H8), MK-9(H8) and MK-9(H6). It has smooth-cylindrical spores which may be formed on the substrate or aerial mycelium as a straight chain to rectiflexible and are non-motile.15–17
The current research demonstrates the isolation, identification, characterization and anti-microbial screening of the Actinomycete strain Streptomyces polyrhachis AS07 from soil along with the production, fermentation and yield optimisation of the secondary metabolic extract. Microbial fermentation was employed to obtain a sufficient yield of the crude metabolite under appropriate growing conditions that facilitate optimal secondary metabolite synthesis. Finally, the results of this study demonstrate the potential biologically active nature of the crude extract and necessitate the need to further examine the hidden potential of this therapeutically significant compound.
Microbial strains used for experimental analysis
A total of ten reference bacterial strains comprising of seven Gram-positive (Staphylococcus aureus ATCC 6538, Bacillus subtilis ATCC 6533, Methicillin-resistant Staphylococcus aureus (MRSA) 13, Propionibacterium acnes strain MTCC 1951, Streptococcus mutans MTCC 890, Staphylococcus warneri MTCC 3050 and Streptococcus pyogenes strain MTCC 1924) and three Gram-negative (Salmonella typhimurium MTCC 3224, Proteus mirabilis ATCC 21100 and Acinetobacter baumannii MTCC 1425) bacteria were used for examining the bioactive potential of the metabolite. Additionally, two reference fungal strains of Candida albicans MCC 1151 and Aspergillus niger MTCC 281 were also tested against the extracted metabolite. The afore-mentioned strains, Staphylococcus aureus ATCC 6538, Bacillus subtilis ATCC 6533 and Proteus mirabilis ATCC 21100 were procured from an authorised distributor for the American Type Culture Collection (ATCC) in India whereas, Streptococcus mutans MTCC 890 and Staphylococcus warneri MTCC 3050 were procured from the Microbial Type Culture Collection and Gene Bank (MTCC), Chandigarh and the rest of the microbial cultures were obtained from existing laboratory test isolates. The active cultures of bacteria were developed by inoculating a loopful of cells from glycerol stock culture to sterile Erlenmeyer flasks containing sterile Luria-Bertani (LB) broth whereas, fungal cultures were grown in Sabouraud Dextrose Broth (SDB) followed by incubation at 37°C for 24 h.
Additionally, an array of biofilm producing bacteria such as Escherichia coli ATCC 8739, Staphylococcus aureus ATCC 6538, Enterococcus faecalis ATCC 29212, Proteus mirabilis ATCC 21100, Bacillus subtilis ATCC 6533, Staphylococcus epidermidis ATCC 12228, Salmonella typhimurium ATCC 14028, Enterobacter aerogenes ATCC 13048 and Shigella boydii ATCC 8700 were procured from an authorised distributor for the American Type Culture Collection (ATCC) in India and Micrococcus luteus MCC 2155, Bacillus cereus MCC 203 were obtained from the National Centre for Cell Science (NCCS), Pune; Klebsiella pneumoniae ESBLCp42 and Pseudomonas aeruginosa PA067 were acquired from existing laboratory isolates. Bacterial cultures were developed by loopful inoculation of cells in a flask containing sterile Luria-Bertani (LB) broth followed by incubation at 37°C for 24 h.
Isolation of streptomyces
A total of 10 soil samples were collected from the Western Ghats of Maharashtra, botanical gardens of B. K. Birla and G.N. Khalsa college campus and few indiscriminate locations in Mumbai, India. The samples were collected from a depth of 10-20 cm using a sterile spatula and each sample was placed in sterile plastic bags and then transferred aseptically to the Microbiology Laboratory at the National Facility for Biopharmaceuticals (NFB) laboratory, India. These samples were sieved, air dried, and then kept at 4˚C for further evaluation. In order to obtain isolated colonies, spread plate technique was employed as performed by Kumar et al.18 Initially, the soil samples were suspended in sterile distilled water (1g/100mL), homogenized by vortexing and 0.1mL of serially diluted sample of 10-6 dilution was spread plated, using a sterile spreader on the surface of International Streptomyces Project 4 (ISP-4) Agar for isolation of Streptomyces. Plates were incubated at 37˚C for 48h. Post incubation period, single distinct colony was selected and maintained by subculturing and was preserved at 4°C.
Antimicrobial screening of isolated actinomycetes
Spot inoculation method was performed on Modified Tryptic Soy Agar (MTSA) plates to conduct preliminary antimicrobial screening of the isolated actinomycetes.11,19 The composition of each MTSA plate consisted of 1g soluble starch, 1.5g pancreatic digest of casein, 0.5g papain digest of soya bean, 0.5g sodium chloride, 0.4g peptone, 0.4g tryptone, 0.3g beef extract, 1g lactose, 0.013g L-cystine, 0.002g bromothymol blue, 1.7g agar, sterile distilled water 100 mL, and pH 6.8. Separate MTSA plates were prepared for seven Gram positive, three-gram Negative bacterial strains and two fungal strains by pour plate method. Using the isolated actinomycete strain, each plate was spot inoculated aseptically. The antibacterial activity was determined by incubating the inoculated plates at 37°C for 24 h, and antifungal activity was determined by incubating the inoculated plates at 30°C for up to 48 h. The antimicrobial activity of the isolate was recorded by observing the inhibition zone around the inoculated spot with the isolated actinomycete strain.
Identification of the actinomycetes
Initially genomic DNA isolation was performed from a 24 h grown actinomycetes strain that displayed antimicrobial activity by the Lysozyme-CTAB method with few modifications.20 After the qualitative analysis of the extracted DNA using agarose gel electrophoresis, the 16S rRNA gene was amplified using Polymerase Chain Reaction in PCR Thermocycler. The isolate was then identified by 16S rRNA gene sequencing as performed by Qureshi et al.,21 with minor modifications. The phylogenetic tree was constructed in MEGA 11 using the Neighbour Joining (NJ) method along with the Bootstrap Phylogeny test with number of bootstrap replications set at 1000 and the Tamura-Nei model.
Metabolite extraction
Parameters such as growing conditions, period of incubation, fermentation medium, and pH were optimized for maximum production of the crude metabolite. In order to develop a potentially growing Streptomyces strain up to 0.5 McFarland Standard in a 250 mL Erlenmeyer flask, the isolated bacterial strain was inoculated in 100 mL ISP-4 medium (with a composition of 10g soluble starch, 1g dipotassium phosphate, 1g magnesium sulphate, 1g NaCl, 2g ammonium sulphate, 2g calcium carbonate, 1mg ferrous sulphate, 1mg manganous chloride and 1mg zinc sulphate)23,24 and incubated for a duration of 5 days at 29°C with a continuous shaking condition set at 160 rpm. Furthermore, the protocol for secondary metabolite extraction was performed in accordance with that as conducted by Leylaie et al.,25 with slight modifications. The bacterial culture obtained was centrifuged at 7000 rpm for 15 min, and cell-free supernatant was harvested and mixed with an equal volume of ethyl acetate and kept in rotary shaker at 160 rpm overnight. The solvent layer was collected and then evaporated at room temperature to obtain yellowish green coloured crude extract. Methanol was used to solubilize the extract and the resulting extract was then refrigerated at 4°C which was later utilised to assess the bioactivity of the extracted metabolite.
GC-MS analysis
Qualitative and quantitative evaluation of the isolated product was performed using qualitatively using Shimadzu GCMS-QP2010 system equipped with Rtx-5MS capillary column (5% diphenyl/ 95% dimethyl polysiloxane) with a length of 30 m, internal diameter of 0.25 mm and film thickness of 0.25 μm.
GC-MS detection analysis was performed using an electron-ionization system with the ionizing energy of 70eV was used. Helium gas (99.999%) was used as the carrier gas at a constant flow rate of 1.51 mL/min and an injection volume of 2μL was employed (20.0 split ratio). The injector as well as the ion-source temperature was set at 200°C. The oven temperature was programmed from 70°C (isothermal for 2 min) to an increase of 300°C for 10 min. In the full scan mode, electron ionization (EI) mass spectra in the range of 40 to 1000 m/z and the total GC running time were 31 min. The relative percent amount of each component was calculated by comparing its average peak area to the total areas and compounds were identified by comparing the mass spectra with library of the National Institute of Standard and Technology (NIST) and the retention indices data obtained from results.26 The mass spectrum of the unknown component was compared with the spectrum of the known components in NIST 08 library.
Determination of anti-microbial property by disc diffusion test
The crude metabolite extract was assessed for its antimicrobial activity against the test microbial strains as performed by Sivasankar et al.,27 with slight modifications. The 24h old bacterial and fungal cultures grown at 30°C in sterile Luria-Bertani (LB) and Sabouraud Dextrose Broth respectively, were initially adjusted to 1.0 × 105 CFU/mL. Later, 2 mL of each suspension culture was spread plated on separate petri dishes containing approximately 20 mL of Luria-Bertani Agar (LA) in case of bacteria and Sabouraud Dextrose Agar (SDA) for fungal cultures. A sterile filter disc (6 mm) impregnated with 1 μL of test sample suspended in methanol was added to each petri plate while, another disc impregnated with a drug used as positive control was placed adjacent to it. Chloramphenicol and Fluconazole each with a concentration of 1mg/mL were used as positive control for bacteria and fungi respectively. The plates spread plated with bacterial cultures were then incubated for 24h at 30°C. On the other hand, the plates spread plated with fungal cultures were incubated for a period of 3-7 days at 30°C. Post the incubation period, the plates were observed for appearance of zone of inhibition.
The Minimum Inhibitory Concentration (MIC) of the microbial strains was estimated by Resazurin Microtiter Assay (REMA) in a flat bottom 96-well clear microtiter plate in accordance with the procedure followed by Elshikh et al.,28 with few variations. Two -fold serial dilution of the methanolic extract was carried out in 1X Mueller Hinton broth from 1000 μg/mL to 1.95 μg/mL. At last, 100 µL volume of microbial culture set at 1.0 × 106 CFU/mL was added to each well and finally, the volume in each well was mixed thoroughly. The microtiter plates were sealed with laboratory grade parafilm and kept for incubation at 37°C for 24h. The MIC of the extracted metabolite was obtained by the addition of 5 µL Resazurin (2 mg/mL) in each well in sterile condition. The plate was again incubated at 37°C for 30 min. Oxidation of Resazurin reagent due to the acidity of the medium resulting from microbial growth gave rise to a colour change of the mixture to pink. The MIC value was defined as the lowest sample concentration indicating no colour change (clear) and exhibited complete inhibition of microorganisms.28
Determination of antioxidant activity by 2,2′-diphenyl-1-picrylhydrazyl Free Radical Scavenging Assay
The strength of the crude metabolite to display antioxidant activity was observed by its ability to scavenge the free DPPH radicals and thereby render the purple-coloured reagent colourless or slightly yellow. Therefore, the crude extract of the bioactive metabolite showing high antimicrobial activity was further studied for its antioxidant activity on the basis of a previously reported protocol by Ser et al.,29 with minor modifications. Streptomyces extract stock with a concentration of 50 µg/mL was prepared and varying extract concentrations ranging from 1 µg/mL to 10 µg/mL were prepared. The total reaction volume was maintained at 4 mL which consisted of sample, methanol as diluent and 2 mL of DPPH reagent was added to each tube. The reaction mixture was incubated in dark for nearly 30-45 min. Following the incubation period, the absorbance was measured at 515 nm using UV Visible spectrophotometer.
The percentage inhibition of the DPPH radical scavenging activity recorded at each concentration was determined by utilising the following formula:
Biofilm inhibition crystal violet assay
According to a procedure performed elsewhere,30 in order to elucidate the anti-biofilm capability of the extracted metabolite, 24h old biofilm producing microbial cultures were initially adjusted to 106 CFU mL-1 in LB broth. Later in a 24-well clear bottom microtiter plate containing 300µL of the microbial suspension, 100µL of crude Streptomyces extract was added aseptically. Additionally, two wells containing 100µL of Chloramphenicol (50mg/mL) and LB broth respectively, along with 300µL of sterile distilled water were treated as positive and negative controls. The plate was incubated for 24h duration at 37°C in static condition to allow microbial growth and maturation. Subsequently, the wells were washed thrice with distilled water to eliminate the free-flowing planktonic cells. Following this step, the plate with remaining adhered biomass was incubated for approximately 30-40 min at room temperature after treating it with of 400µL of 1% Crystal Violet solution. The plates were then drained and rinsed three times with distilled water to remove unbound dye. After the washing step, the plates were dried in the hot-air oven at 40°C for 10-20 min. Once dried, 400μL of methanol was added in each well. Absorbance was measured at 570nm wavelength in ELISA plate reader and percentage inhibition was calculated by the formula:
Qualitative and quantitative anti-quorum sensing assay
For this experimental study, a wild strain of Chromobacterium violaceum was used as a reporter strain for screening of anti-quorum activity of the soil isolate using disc diffusion method as previously reported by Miao et al.31 Firstly, the reporter strain was inoculated in LB broth and incubated for 24h at 37˚C. The C. violaceum suspension was adjusted at 10-6 CFU mL-1 using Luria Bertani broth and 100µL of the resulting suspension was spread plated on sterile Nutrient Agar plate. On this plate, a sterile filter disc (6mm) impregnated with 10µL of the extracted metabolite and another disc consisting of 10µL of Chloramphenicol (50 μg/mL) were placed. The plate was then incubated for a period of 24h at 37˚C.
Motility assay
To observe the effect of Streptomyces metabolite on swimming and swarming motility of the gram-negative bacterium Pseudomonas aeruginosa PA01, the motility assay was conducted as performed by O’May et al.,32 in glass petri plates consisting of Luria Bertani Agar uniformly mixed with 100μL of the methanolic extract. To observe the swimming motion of the bacterium in presence of the metabolite, 5μL of 24h old bacterial culture adjusted at 108 CFU mL-1 was placed directly into the centre of the plate comprising of semi-solid Luria Bertani Agar evenly mixed with 100μL of the methanolic extract. On the other hand, to observe the swarming motility of the test bacteria in the presence of the extracted sample, a volume of 5μL of the same bacterial suspension used previously for swimming motility was placed directly on the agar surface. Control plates for both the motility types consisted of evenly mixed LB agar with a small volume of methanol with test bacterium placed in the centre. All the plates were incubated in upright position at 37°C for 24h duration and post the incubation period, the diameter of the motility zones was measured, and percent motility was recorded.
Statistical analysis
All the experimental procedures involved in determination of anti-microbial, antioxidant and anti-biofilm activity of the crude extract were performed thrice. The resultant data was analysed using statistical software and the values were represented as Mean ± Standard Deviation. Wherever applicable the data was subjected to one-way Analysis of Variance (ANOVA) test. The data was considered to be statistically significant at p < 0.05.
Isolation and preliminary antimicrobial screening of the isolated actinomycetes strain
The actinomycetes strain, namely AS07 was successfully isolated from the ten soil samples by obtaining pure culture through growth on ISP-4 medium. The isolated strain exhibited considerable zone of inhibition when tested against all the test organisms. Thus, it was selected further for experimental analysis.
Identification of the As07 strain
The result of the isolated strain obtained by next generation 16S rRNA sequencing analysis was used for comparison against all available sequences in GenBank (NCBI, USA), depicting that AS07 was closely related to the genus Streptomyces. After performing phylogenetic analysis, the results indicated that the AS07 strain showed highest similarity against Streptomyces cadmiisoli ZFG47 strain (83 %). The neighbour-joining phylogenetic tree of Streptomyces polyrhachis AS07 generated by MEGA 11 (Figure 1) indicated that the isolated Streptomyces strain belonged to genus Streptomyces. The AS07, 16S rRNA gene sequence was submitted to NCBI GenBank with accession number OL343679.1.
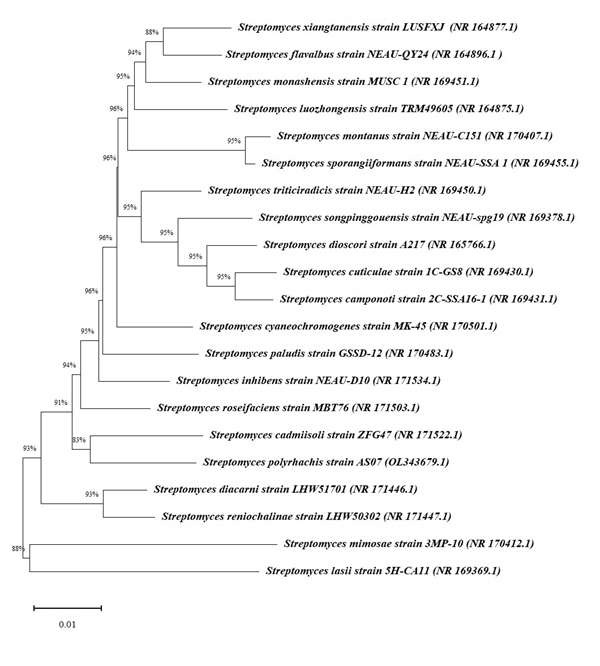
Figure 1 Phylogenetic tree of AS07 strain obtained by Neighbour-joining (NJ) method using MEGA 11 software. The branch node number show percent bootstrap support. The accession numbers of the organisms are included in parentheses and the bar scale value 0.01 indicate the nucleotide substitutions per site.
GC–MS spectroscopy
Identification of several constituents present in the extracted metabolite was conducted by GC-MS technique. The resultant GC-MS chromatogram of the secondary crude metabolite consisted of 28 peaks with their respective retention time in minutes and peak area. After performing comparison of the obtained data with that from NIST database, the closest hit for each peak was determined (Figure 2). The analysis identified a range of compounds varying from different types of azoles, monocarboxylic acids, secondary alcohols, phenolic compounds, and other organic components (Table 1). Additionally, because of the proportional relationship between peak area and constituent quantity, the constituent present in the sample bearing highest portion of the crude extract was determined by its peak area percent. The data obtained reported Cyclo(Phe-Pro) (36.44%) as the most prevalent component followed by Gancidin W (30.13%); 2-Decene, 3-methyl-, (Z)- (12.29%); Cyclo(Phe-Val) (3.41%); 2(3H)-Furanone, dihydro-5-(hydroxymethyl)-3,3-dimethyl- (2.35%); Heptadecylamine (1.84%); 2-Amino-4-methyl-5-acetylthiazole (1.72%); Benzene acetic acid (1.66%); 1-(phenylacetyl)piperazine (1.5%); 1-Methyl-2-pyrrolidone-4-carboxamide (1.16%) and few other components detected in minor quantities. The aforementioned compounds had a retention time of 41.54, 36.237, 15.398, 19.963, 19.714, 15.93, 15.875, 8.204, 20.714, 14.115 minutes respectively. Amongst the 28 compounds detected, 2 compounds were earlier reported for their antimicrobial and anticancer activity (1-Phenyl-5-[(2',3',4,4'-tetramethoxy[1,1'-biphenyl]-2-yl)oxy]-1H-tetraazole,33 2-Amino-4-methyl-5-acetylthiazole)34 and one compound (1-Methyl-2-pyrrolidone-4-carboxamide)35 was studied earlier for its antimicrobial potential; other 2 compounds were investigated in the past for their antifungal property (Propanoic acid36 and Palmitic acid);37 1 compound (3,5-bis(1,1-dimethylethyl)-)38 was reported previously for its antioxidant and anti-cancer property. None of these compounds have not been reported to be isolated previously from Streptomyces sp.
|
Sr. No. |
Compound name |
Molecular formula |
MW (g/mol) |
Retention time (minutes) |
% Area |
|
1 |
1-Phenyl-5-[(2',3',4,4'-tetramethoxy[1,1'-biphenyl]-2-yl) oxy]-1H-tetraazole |
C23H22N4O5 |
434.4 |
5.75 |
0.28 |
|
2 |
Propanoic acid |
C26H24N2O8S4 |
620.7 |
8.1 |
0.29 |
|
3 |
Benzene acetic acid |
C20H20O5 |
340.4 |
8.204 |
1.66 |
|
4 |
2-Hexanol |
C8H18O2S |
178.29 |
8.267 |
0.17 |
|
5 |
3,5-bis(1,1-dimethylethyl)- |
C14H22O |
206.32 |
11.546 |
0.2 |
|
6 |
1-Methyl-2-pyrrolidone-4-carboxamide |
C6H10N2O2 |
142.16 |
14.115 |
1.16 |
|
7 |
Palmitic acid |
C16H32O2 |
256.42 |
14.697 |
0.69 |
|
8 |
2-Decene, 3-methyl-, (Z)- |
C11H22 |
154.29 |
15.398 |
12.29 |
|
9 |
2-Amino-4-methyl-5-acetylthiazole |
C6H8N2OS |
156.21 |
15.875 |
1.72 |
|
10 |
Heptadecylamine |
C17H37N |
255.5 |
15.93 |
1.84 |
|
11 |
1-Pentanamine, N-nitro-N-pentyl- |
C10H22N2O2 |
202.29 |
15.983 |
0.2 |
|
12 |
cis-1-Ethyl-2-Methylcyclopentane |
C8H16 |
112.21 |
16.95 |
0.25 |
|
13 |
(3Z)-1,3,13-Tetradecatriene |
C14H24 |
192.34 |
18.418 |
0.28 |
|
14 |
1-Deoxyhexitol |
C6H14O5 |
166.17 |
18.711 |
0.05 |
|
15 |
3-Heptene, 4-methoxy- |
C8H16O |
128.21 |
18.754 |
0.18 |
|
16 |
5-Acetyl-2-methylpyridine |
C8H9NO |
135.16 |
18.925 |
0.53 |
|
17 |
Cyclo(Ala-Phe) |
C12H14N2O2 |
218.25 |
18.959 |
0.8 |
|
18 |
2-Isobutyl-4-methyl-1,3-dioxolane |
C8H16O2 |
144.21 |
19.05 |
0.07 |
|
19 |
2(3H)-Furanone, dihydro-5-(hydroxymethyl)-3,3-dimethyl- |
C7H12O3 |
144.17 |
19.714 |
2.35 |
|
20 |
Cyclo(Phe-Val) |
C14H18N2O2 |
246.3 |
19.963 |
3.41 |
|
21 |
1-(phenylacetyl)piperazine |
C12H16N2O |
204.27 |
20.714 |
1.5 |
|
22 |
8-Chlorooctyl ethyl carbonate |
C11H21ClO3 |
236.73 |
21.05 |
0.11 |
|
23 |
4-Methyl-1-phenyl-3-penten-1-ol |
C12H16O |
176.25 |
23.467 |
0.57 |
|
24 |
Thiazolo[2,3-c] [1,2,4] triazole, 6-methyl-3-(O-tolyloxymethyl)-5,6-dihydro- |
C13H15N3OS |
261.34 |
23.733 |
0.84 |
|
25 |
Oleamide |
C18H35NO |
281.5 |
23.798 |
0.87 |
|
26 |
Dibenzyl-3,5-piperazine |
C18H18N2O2 |
294.3 |
24.28 |
1.11 |
|
27 |
Gancidin W |
C11H18N2O2 |
210.27 |
36.237 |
30.13 |
|
28 |
Cyclo(Phe-Pro) |
C14H16N2O2 |
244.29 |
41.54 |
36.44 |
Table 1 Constituents present in crude metabolic extract identified by GC-MS technique
Antimicrobial analysis by agar diffusion method
To evaluate the nature of the crude extract as a potential antimicrobial compound, the secondary metabolic extract was treated against a range of microbial strains. The metabolite extract obtained from the isolate Streptomyces polyrhachis depicted significant antibacterial activity against Staphylococcus aureus (20.96 ± 0.04 mm) followed by Streptococcus pyogenes (17.93 ± 0.06 mm) and Bacillus subtilis (16.97 ± 0.06 mm), whereas highest anti-fungal activity was observed against Aspergillus niger (16.92 ± 0.06 mm) (Table 2) (Figure 3). The resultant antimicrobial activity probably might have been observed due to the presence of antimicrobial agents in the test sample as confirmed earlier by GC-MS analysis. Compounds such as 1-Phenyl-5-[(2',3',4,4'-tetramethoxy[1,1'-biphenyl]-2-yl) oxy]-1H-tetraazole,33 1-Methyl-2-pyrrolidone-4-carboxamide,34 2-Amino-4-methyl-5-acetylthiazole35 and Propanoic acid,36 Palmitic acid37 obtained in the extracted metabolite have been reported previously as potent anti-microbial and anti-fungal agents respectively. Therefore, due to the abundant presence of antimicrobial compounds in the secondary metabolite obtained from the Streptomyces sp., a considerable antimicrobial activity was observed against the test bacterial and fungal strains.
|
Sr. No. |
Microbial strains |
Inhibition zone (mm) |
|
|
Metabolic extracta |
Positive controla |
||
|
1 |
Staphylococcus aureus |
20.96 ± 0.04 |
32.02 ± 0.06 |
|
2 |
Bacillus subtilis |
16.97 ± 0.06 |
27.98 ± 0.02 |
|
3 |
MRSA |
12.93 ± 0.05 |
26.98 ± 0.02 |
|
4 |
Propionibacterium acnes |
15.91 ± 0.06 |
27.97 ± 0.05 |
|
5 |
Salmonella typhimurium |
14.92 ± 0.06 |
25.05 ± 0.04 |
|
6 |
Streptococcus mutans |
14.92 ± 0.06 |
29.04 ± 0.04 |
|
7 |
Proteus mirabilis |
15.93 ± 0.05 |
31.50 ± 0.04 |
|
8 |
Staphylococcus warneri |
14.94 ± 0.07 |
29.08 ± 0.08 |
|
9 |
Streptococcus pyogenes |
17.93 ± 0.06 |
28 ± 0 |
|
10 |
Acinetobacter baumannii |
16.91 ± 0.07 |
29.99 ± 0.01 |
|
11 |
Candida albicans |
14.90 ± 0.07 |
20.99 ± 0.02 |
|
12 |
Aspergillus niger |
16.92 ± 0.06 |
31.99 ± 0.01 |
Table 2 Antimicrobial activity of the metabolic extract produced by Streptomyces polyrhachis
aValues are mentioned as Mean ± Standard Deviation (n=3).
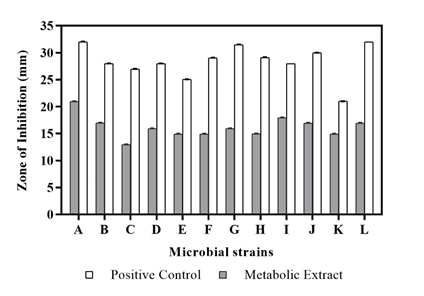
Figure 3 Anti-microbial Activity of metabolic extract from Streptomyces polyrhachis against (A) Staphylococcus aureus, (B) Bacillus subtilis, (C) MRSA, (D) Propionibacterium acnes, (E) Salmonella typhimurium, (F) Streptococcus mutans, (G) Proteus mirabilis, (H) Staphylococcus warneri, (I) Streptococcus pyogenes, (J) Acinetobacter baumannii, (K) Candida albicans, (L) Aspergillus niger.
Antimicrobial analysis by broth dilution method
MIC (Minimum Inhibitory Concentration) of the metabolic extract was determined against all test organisms by 2-fold serial broth dilution procedure (Figure 4). The crude extract efficiently restricted the growth of Staphylococcus aureus at a MIC value of 41.67 ± 14.73 μg/mL followed by Candida albicans at an inhibitory concentration of 62.47 ± 29.46 μg/mL. On the other hand, the highest minimum inhibitory concentration of the metabolite was observed in the cases of Streptococcus pyogenes (666.67 ± 235.70 μg/mL) and Staphylococcus warneri (416.67 ± 117.85 μg/mL) (Table 3). These values indicate the higher resistance ability of these organisms against the test sample. It can also be said that the ability of the metabolite extracted from Streptomyces species to exhibit considerable antimicrobial activity might have resulted due to the existence of antimicrobial components identified by GC-MS analysis.
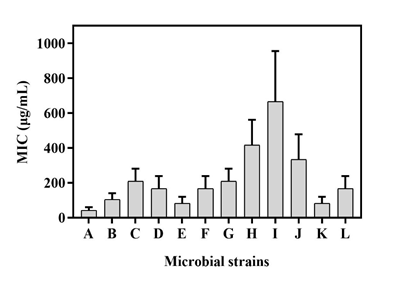
Figure 4 MIC value of metabolic extract from Streptomyces polyrhachis against (A) Staphylococcus aureus, (B) Bacillus subtilis, (C) MRSA, (D) Propionibacterium acnes, (E) Salmonella typhimurium, (F) Streptococcus mutans, (G) Proteus mirabilis, (H) Staphylococcus warneri, (I) Streptococcus pyogenes, (J) Acinetobacter baumannii, (K) Candida albicans, (L) Aspergillus niger.
|
Sr. No. |
Microbial strains |
Minimum inhibitory concentrationa (MIC) (μg/mL) |
|
1 |
Staphylococcus aureus |
41.67 ± 14.73 |
|
2 |
Bacillus subtilis |
104.17 ± 29.46 |
|
3 |
MRSA |
208.33 ± 58.93 |
|
4 |
Propionibacterium acnes |
166.67 ± 58.93 |
|
5 |
Salmonella typhimurium |
83.33 ± 29.46 |
|
6 |
Streptococcus mutans |
166.67 ± 58.93 |
|
7 |
Proteus mirabilis |
208.33 ± 58.93 |
|
8 |
Staphylococcus warneri |
416.67 ± 117.85 |
|
9 |
Streptococcus pyogenes |
666.67 ± 235.70 |
|
10 |
Acinetobacter baumannii |
333.33 ± 117.85 |
|
11 |
Candida albicans |
62.47 ± 29.46 |
|
12 |
Aspergillus niger |
166.67 ± 58.93 |
Table 3 Minimum Inhibitory Concentration of metabolic extract against test microbial strains
aValues are mentioned as Mean ± Standard Deviation (n=3).
Antioxidant activity analysis by DPPH free radical scavenging assay
The scavenging of free radicals of DPPH was observed by decrease in absorbance at 517nm induced by the secondary metabolic extract produced by Streptomyces species (Table 4). The data obtained for antioxidant activity in case of both Ascorbic acid and metabolic extract had an IC50 value of 5.85μg/mL and 5.50μg/mL respectively (Figure 5). Although the metabolic extract exhibited a decrease in percent scavenging activity with increasing concentration as compared to ascorbic acid, it had an IC50 value approximately the same as ascorbic acid. Moreover, the observed antioxidant activity of the crude metabolite might have been observed due to the presence of 3,5-bis(1,1-dimethylethyl). The relatively lower antioxidant activity of the metabolic extract must have resulted due to lower concentration of this antioxidant component in the test sample. Also, a previous study in the past undertaken by Dai et al has reported this component to be an effective anti-oxidant biomolecule.38
|
Concentration (µg/mL) |
Ascorbic acid (ASB) |
Metabolic extract (ME) |
||
|
Absorbance (515 nm) |
% Free radical scavenging activitya |
Absorbance (515 nm) |
% Free radical scavenging activitya |
|
|
Control (DPPH) |
0.316 |
|
0.316 |
|
|
1 |
0.31 |
1.90 ± 0.02 |
0.313 |
0.95 ± 0.01 |
|
2 |
0.309 |
2.22 ± 0.02 |
0.31 |
1.90 ± 0.01 |
|
3 |
0.3 |
5.06 ± 0.07 |
0.307 |
2.85 ± 0.03 |
|
4 |
0.27 |
14.56 ± 0.02 |
0.28 |
11.39 ± 0.03 |
|
5 |
0.22 |
30.38 ± 0.02 |
0.245 |
22.47 ± 0.01 |
|
6 |
0.15 |
43.04 ± 0.03 |
0.192 |
39.24 ± 0.03 |
|
7 |
0.12 |
62.03 ± 0.02 |
0.163 |
48.42 ± 0.02 |
|
8 |
0.1 |
68.35 ± 0.05 |
0.158 |
50.00 ± 0.04 |
|
9 |
0.07 |
77.85 ± 0.04 |
0.149 |
52.85 ± 0.05 |
|
10 |
0.03 |
90.51 ± 0.04 |
0.121 |
61.71 ± 0.01 |
Table 4 Percent Free Radical Scavenging Activity of Streptomyces extract from 1 µg/mL to 10 µg/mL
a% Free Radical Scavenging Activity is expressed as Mean ± Standard Deviation (n=3).
Effect of crude metabolic extract on biofilm development
The anti-biofilm activity of the metabolite extract of the isolate was tested against a wide range of biofilm producing organisms (Figure 6). The extracted sample displayed promising antibiofilm effect against test biofilm synthesizing organisms. The metabolite showed highest biofilm inhibition against Bacillus subtilis (80.40 ± 0.05 %) followed by Escherichia coli (75 ± 0.06 %) and Bacillus cereus (74.50 ± 0.05 %) (Table 5). The observed anti-biofilm activity of the extract was obtained owing to the presence of several antimicrobial constituents present in the crude metabolite extracted from Streptomyces polyrhachis. A recent investigation on natural anti-biofilm agents has reported antimicrobial compounds to be potent anti-biofilm agents which can exhibit a broad range of mechanisms across the microbial species and affect the biofilm formation process thereby paving a way towards the development of therapeutic strategies against various pathogens.39

Figure 6 Percent biofilm inhibition of the extract against (A) Pseudomonas aeruginosa, (B) Klebsiella pneumoniae, (C) Escherichia coli, (D) Enterococcus faecalis, (E) Proteus mirabilis, (F) Bacillus subtilis, (G) Micrococcus luteus, (H) Bacillus cereus, (I) Staphylococcus aureus, (J) Salmonella typhimurium, (K) Enterobacter aerogenes, (L) Shigella boydii, (M) Staphylococcus epidermidis.
|
Sr.No |
Biofilm producing organisms |
% Inhibitiona of metabolic extract obtained from Streptomyces polyrhachis |
|
1 |
Pseudomonas aeruginosa |
64.80 ± 0.08 |
|
2 |
Klebsiella pneumoniae |
67.00 ± 0.01 |
|
3 |
Escherichia coli |
75.00 ± 0.06 |
|
4 |
Enterococcus faecalis |
50.00 ± 0.04 |
|
5 |
Proteus mirabilis |
39.30 ± 0.05 |
|
6 |
Bacillus subtilis |
80.40 ± 0.05 |
|
7 |
Micrococcus luteus |
70.60 ± 0.06 |
|
8 |
Bacillus cereus |
74.50 ± 0.05 |
|
9 |
Staphylococcus aureus |
58.20 ± 0.07 |
|
10 |
Salmonella typhimurium |
69.80 ± 0.02 |
|
11 |
Enterobacter aerogenes |
64.60 ± 0.06 |
|
12 |
Shigella boydii |
67.50 ± 0.05 |
|
13 |
Staphylococcus epidermidis |
66.00 ± 0.06 |
Table 5 Percent inhibition for anti-biofilm activity of the extract against 13 actively biofilm synthesizing bacterial strains
aValues are expressed as Mean ± Standard Deviation (n=3).
Assessment of anti-quorum sensing
Production of purple coloured violacein pigmentation by quorum sensing communication in Chromobacterium violaceum provides a naturally occurring and readily recognizable phenotype which helps in the evaluation of anti-QS compounds in test sample. Metabolite extract of Streptomyces polyrhachis displayed 46.42 % inhibition of violacein as mentioned in Table 6 and displayed in Figure 7. This property of the test sample to inhibit the quorum sensing ability of C. violaceum can be attributed to the presence of anti-microbial compounds present in the secondary metabolite extract obtained from the isolated Streptomyces strain.
|
Test Organism |
Zone of inhibitiona (mm) |
Percent Inhibition |
|
|
Metabolic extract |
Positive Control (Chloramphenicol) |
||
|
Chromobacterium violaceum |
14.95 ± 0.06 |
27.97 ± 0.05 |
46.42 |
Table 6 Anti-QS activity of metabolic extract from Streptomyces polyrhachis against the biosensor strain Chromobacterium violaceum by disc diffusion method
aValues are expressed as Mean ± Standard Deviation (n=3). polyrhachis against the biosensor strain Chromobacterium violaceum by disc diffusion method.
Motility assay
For motility assay, the metabolite extract was tested against Pseudomonas aeruginosa PA01 strain. The larger the diameter of the zone, less effective is the metabolite to inhibit the movement of the bacteria and vice-versa. Pseudomonas aeruginosa showed 5mm diameter of swimming zone, indicating 50% of swimming ability of bacterium in presence of the metabolite extracted from soil bacteria Streptomyces polyrhachis (Figure 8). Additionally, the test bacterium had a 75mm diameter of swarming zone, indicating nearly 16% of swarming ability of the organism in presence of the metabolite extracted from soil bacteria Streptomyces polyrhachis as mentioned in Table 7.
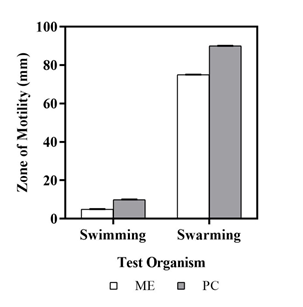
Figure 8 Zone of swimming and swarming of Pseudomonas aeruginosa when treated with metabolic extract.
|
Test organism |
Swimming Assay |
Swarming Assay |
||||
|
Zone of swimminga (mm) |
Percent Swimming |
Zone of Swarminga (mm) |
Percent Swarming |
|||
|
Metabolic extract |
Positive control |
Metabolic extract |
Positive control |
|||
|
Pseudomonas aeruginosa |
4.93 ± 0.06 |
9.95 ± 0.07 |
50 |
74.95 ± 0.07 |
89.95 ± 0.04 |
16.67 |
Table 7 Motility ability of Pseudomonas aeruginosa in presence of metabolic extract from S. polyrhachis
aValues are expressed as Mean ± Standard Deviation (n=3).
In our study, initially Streptomyces strain was isolated from soil followed by next generation 16S rRNA sequencing and phylogenetic analysis, thereby confirming that the isolated strain belonged to Streptomyces sp. The metabolites produced by Streptomyces polyrhachis had shown appreciable antimicrobial activity against a range of said organisms, which suggested that it could be a potent source of effective bioactive molecules against challenging pathogenic microorganisms. In addition, the metabolite also came across as a potent antioxidant agent, showed considerable anti-quorum sensing capability and also acted as an efficient inhibitor for biofilm synthesis.
Ethics approval and consent to participate: Not applicable.
Consent for publication: Not applicable.
Data availability statement: All data generated or analysed during this study are included in this published article.
Ethical statement: No animals were harmed during this study.
Additional Information: Organism NCBI Accession Number: OL343679.1
Streptomyces polyrhachis strain AS07 16S ribosomal RNA gene.
None.
The authors state that there is no conflict of interest.
None.

©2022 Jha, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.