Journal of
eISSN: 2572-8466


Research Article Volume 8 Issue 1
1Research-Development Department, Center for Genetic Engineering and Biotechnology of Sancti Spiritus, Cuba
2Clinical Research Direction, Center for Genetic Engineering and Biotechnology, Cuba
3Quality Control Direction, Center for Genetic Engineering and Biotechnology, Cuba
Correspondence: Maylin Pérez-Bernal, Center for Genetic Engineering and Biotechnology of Sancti Spiritus, Cuba
Received: January 11, 2021 | Published: February 19, 2021
Citation: Pérez-Bernal M, Hernández C, Delgado M, et al. Full validation of an ELISA for quantification of antibodies against Haemophilus influenzae type b capsular polysaccharide. J Appl Biotechnol Bioeng. 2021;8(1):24-31 DOI: 10.15406/jabb.2021.08.00247
Since 1981, ELISA methods have been used for the quantification of antibodies against Haemophilus influenzae type b (Hib), but their complete validation has been never reported. This work described the full validation of an ELISA for the quantification of antibodies against Hib capsular polysaccharide, following eleven parameters recommended by European Medicines Agency and FDA guidelines. The method satisfied the acceptance criteria for the required parameters. The use of percent of inhibition and parallelism fulfillment as acceptance criteria for specificity was discussed. This ELISA will be a reliable device for the evaluation of Hib-vaccination efficacious.
Keywords: accuracy, anti-PRP antibodies, calibration curve, precision
PRP, polyribosylribitol-phosphate; ELISA, enzyme-linked immunosorbent assay; MRD, minimum required dilution
Haemophilus influenzae type b (Hib) is a cause of invasive infections such as meningitis, pneumonia, sepsis, and epiglottitis in children. In developing countries, the incidence of Hib disease is higher than in industrialized countries. It occurs among children younger than five years of age, and approximately two-thirds of all cases among children younger than 18 months of age.1 Hib is particularly virulent because its capsule contains a repeating polymer of ribosyl and ribitol phosphate (polyribosylribitol-phosphate, PRP), which enables the organism to evade complement-mediated killing and avoid splenic clearance.2 Like other polysaccharides, the PRP of the Hib capsule is a T-independent antigen and not immunogenic when administered as a vaccine in infancy.3 Access to synthetic complex carbohydrate-based vaccines is feasible and provides a strategy in the fight against Hib infections.4 The quantification of antibodies produced after vaccination is necessary to confirm whether it has been successful. Short- and long-term protection against Hib infections is achieved by anti-PRP antibody concentrations of 0.15 and 1.0 mg/mL, respectively.5
Serological analysis by Enzyme-Linked Immunosorbent Assay (ELISA) has been widely used for evaluation of the immune response to vaccines. Since 1981, some ELISA methods have been reported for the quantification of antibodies against Hib capsular polysaccharide in serum,6,7 but none of them has described the complete validation of the method, as recommended by the current regulatory guidelines. Full validation refers to the establishment of all validation parameters that apply to sample analysis for the bioanalytical method.8 The aim of this study was the full validation of an indirect ELISA for the quantification of anti-PRP antibodies in serum of vaccinated patients, to prove the immunogenicity of a conjugated vaccine with a PRP-synthetic antigen. The validation was based on eleven parameters; most of them were described by the European Medicines Agency (EMA) guideline.9 The preparation of quality controls and the robustness of the ELISA were validated through FDA guidance.10
Reagents
The PRP-synthetic antigen, conjugated to human albumin (PRP-HA, CIGB, Cuba), was used for ELISA plate coating. This antigen and the PRP-natural antigen, conjugated to human albumin (HbOHA, NIBSC, UK), were used in the inhibition assay. The human anti-Haemophilus influenzae b reference serum (NIBSC, UK), with 69.4µg/mL of IgG anti-PRP, was used to prepare the calibration curve. For the evaluation of dilutional linearity, it was used a standard provided by the Tropical Medicine Institute (IPK, Havana, Cuba) with 101.5µg/mL of IgG anti-PRP.
Serum samples
Individual serum samples from 60 children included in the clinical assay “Evaluation of safety and immunogenicity of liquid pentavalent vaccine DPT-HB-HIB, compared with HEBERPENTA® vaccine administration in healthy children, schedule 2-4-6 months. Code: IG/VHI/DPT-HB-Hib/06-01.”
Enzyme-linked immunosorbent assay
Polystyrene 96-well microtiter plates (Thermo Scientific™ Nunc™ Maxisorp), with strip plate modules, were coated with 1.0 μg/mL PRP-HA in 100μL/well coating buffer (phosphate-buffered saline, PBS, pH 7.2-7.4) and incubated 90 min at 37°C. Wells were washed three times with 380μL/well washing buffer (PBS and 0.05% Tween-20) and blocked with 350μL/well blocking buffer (PBS, 1% bovine serum albumin and 0.1% Tween-20) 1 h at 37ºC. The plate was washed once, and 100 μL of samples, diluted with dilution buffer, (DB: PBS, 1% bovine serum albumin, 0.1% Tween-20 and 10mM EDTA), were added to wells and incubated 90 min at (22-25) °C. Plates were washed three times and 100μL/well of peroxidase (HRP)-conjugated goat antihuman IgG in rabbit (Dako, Denmark; 1:5000 in DB) were added and incubated 1 h at 37ºC. After three washes, 100μL/well o-phenylenediamine dihydrochloride (OPD, 0.6 mg/mL)) with 0.015% hydrogen peroxide in the citrate-phosphate buffer, pH 5.0, were added and incubated 20 min at (22-25)°C in the dark. Color development was stopped by adding 100μL/well 2.66 M HCl. Absorbance was measured at 492nm (A492nm) in a microplate reader (Labsystems Multiskan® Plus, Finland).
Calibration curve
The calibration curve was based on eight serial two-fold dilutions of NIBSC standard with DB, from 250 to 1.95 ng/mL. It was repeated in triplicated during six days by two analysts, three days each one. For the evaluation of the intra-curve repeatability, the accuracy of 75% of the points of the curve, assessed for percent of relative error (%RE), should be less than 20%, except at the lower point of the curve, which should be less than 25%. The precision and accuracy inter-curves were assessed by the coefficient of variation expressed as a percentage (%CV) and %RE, respectively. Both should be less than 15%, except at the lower point of the curve, which should be less than 20%.
%RE and %CV were determined by the formulas:
%RE= (Mean of back-calculated concentration – Nominal concentration) x 100/ Nominal concent.
%CV= Standard deviation x 100/ Mean of back-calculated concentration
Minimum required dilution (MRD)
A pool of ten serum samples, with a low concentration of anti-PRP antibodies, was spiked with NIBSC standard to obtain a concentration of 250 ng/mL of anti-PRP antibodies. The pool was serially two-fold diluted with DB, from 250 ng/mL to 1.95 ng/mL. The percent of recovery (%R) was calculated for each dilution point, using the back-calculated concentration of the pool in respect to the nominal concentration:
%R = (Back-calculated concentration/Nominal concentration) x 100
The minimum dilution with a percent of recovery between 80 and 120% was selected as MRD.
Specificity
The evaluation of specificity was performed by an inhibition assay. High (125ng/mL) and low (5ng/mL) concentrations of NIBSC standard, diluted at MRD in a matrix with low anti-PRP concentration, were pre-incubated independently with 200μg/mL of PRP-HA and with 200μg/mL of HbOHA. The non-inhibited control contained PBS instead of the antigen. After 1h at (22-25)ºC, the inhibited and non-inhibited specimens were added into the plate wells, which were coated previously with 1.0 mg/mL PRP-HA and blocked with blocking buffer. The assay had three replications. The percent of inhibition (%I) was calculated as follows:
%I = 100 – [(mean A492nm inhibited) / (mean A492nm non-inhibited)] x 100
ELISA was considered specific if the percent of inhibition was higher than 90%.
Selectivity
Selectivity was evaluated using two types of samples: ten individual hemolytic sera and ten individual lipemic sera. First, it was necessary to quantify the endogenous anti-PRP concentration in each sample. After that, samples were spiked with NIBSC standard to prepare pools with low anti-PRP concentration (between 5 and 8 ng/mL). Samples were duplicated into the ELISA plate. The percent of recovery (%R) was calculated by the formula:
Back-calculated concentration for the pool
Sample was not interfering if the percent of recovery was between 80 and 120%.
Accuracy and precision
A pool of ten serum samples with low anti-PRP concentration, used at MRD, was spiked with NIBSC standard to obtain five samples (S1 to S5) with anti-PRP concentrations into the calibration range as follows:
S1: Concentration corresponding with low quantification limit (2 ng/mL)
S2: Concentration three-fold high than S1 (6 ng/mL)
S3: Midpoint of the calibration curve (25 ng/mL)
S4: High concentration of the calibration curve (125 ng/mL)
S5: Concentration corresponding with high quantification limit (250 ng/mL)
Spiked pools of sera were duplicated in two positions into the ELISA plate. Accuracy and precision were evaluated by repeating the assay in six analytical runs by two analysts. The standard deviation was estimated by a variance component analysis, using a restricted maximum likelihood method within the framework of a nested ANOVA.
The intra- and inter-assays precision were expressed by the %CV and it was calculated by the formulas:
%CV intra-assay = (√variance of error) x 100/ mean of back-calculated concentration
%CV inter-assays = (√∑variance components) x 100/ mean of back-calculated concentration
Intra- and inter-assays precision at each anti-PRP concentration should not exceed 20%, except at the S1 and S5, where it should not exceed 25%. The intra- and inter-assays accuracy (%RE) at each anti-PRP concentration level should be less than 20%, except at the S1 and S5, where it should be less than 25%.
Quantification range
A standard curve was prepared using as diluent a pooled serum with a low concentration of anti-PRP, diluted with DB at MRD. Experiments were performed in six ELISAs by two analysts, and the curve was analyzed in duplicates. Quantification range was established between the lowest and the highest concentration values for which the precision (%CV) and accuracy (%RE) are both ≤20%.
Dilutional linearity
Samples exceeding the upper limit of the quantification of the calibration curve were used for the assessment of dilutional linearity. A curve was prepared with serial two-fold dilutions of the IPK standard, using as diluent a pooled serum with a low concentration of anti-PRP (0.6 ng/mL), diluted with DB at MRD. The curve had thirteen dilution points, with a nominal concentration range from 2000 to 1.088 ng/mL. The back-calculated concentration for each diluted sample should be within 20% of the nominal value. The back-calculated concentration in serum was calculated by correcting the back-calculated concentration for the dilution factor. The precision of data (%CV) from back calculated concentration in serum, through the quantification range, should be ≤20%.
Parallelism
Seven pools of samples with high anti-PRP concentration were diluted serially 1:2 with a pooled serum with a low concentration of anti-PRP, diluted with DB at MRD. Pool dilutions were included in the calibration range. Data precision (%CV) between sample pools in a dilution series should not exceed 30%.
Quality controls
Quality controls (QCs) were prepared with a pool of serum samples with high anti-PRP concentration, diluted in DB. Three anti-PRP concentrations within the calibration range were established: low-level (LQC) near to the lower point of the calibration curve, mid-level (MQC) in the midrange of the curve, and high-level (HQC) near to the higher point of the curve. Each QC was duplicated into the ELISA plate and they were quantified in six ELISAs by two analysts. For the definition of the nominal anti-PRP concentration of QCs, the mean concentration of each one with data from the six assays was calculated, with a precision of ≤20%. The accuracy intervals of QCs were defined within±20% of their nominal concentrations.
Robustness
There were made small changes in critical parameters of the ELISA procedure: variation in±0.2 mg/mL the OPD concentration; use of bovine serum albumin from four suppliers (AppliChem, Sigma, Spectrum, and Roche) for the preparation of blocking and dilution buffers; use of Costar™ 3590 High Binding ELISA plate instead of Nunc™ MaxiSorp strip plate module; use of a different lot of horseradish peroxidase (HRP)-conjugated goat antihuman IgG in rabbit (Dako, Denmark). Each treatment was evaluated in triplicates. The robustness was proved by the fulfilling of the QCs accuracy intervals.
Stability of samples and quality controls
The origin of samples and QCs was the same, for that reason the stability of both was proved by the fulfilling of the QCs accuracy intervals. Short-term stability (6 hours at room temperature (20-25)ºC and 6 days at (2-8)ºC) and freeze–thaw stability (three and five freeze-thaw cycles) were tested. Data were measured in triplicates. For the evaluation of freeze–thaw stability, aliquots of each QC were thawed unassisted at room temperature. When completely thawed, the aliquots were refrozen at -20ºC for at least 20 hours.
Statistical analysis
Statistical analyses were performed using Microsoft® Office Excel (2010) and the Statistical Package for Social Science 15.0. Curve fitting was achieved using Sigma Plot 12.
Calibration curve
Calibration curve concentration-response data from a minimum of three independent runs should be analyzed when establishing a calibration model.11 In the present study, the calibration model was achieved with data obtained from six independent runs. Figure 1 shows the average of six calibration curves, each prepared with the NIBSC reference serum, spanning the anti-PRP concentration range from 1.95 to 250 ng/mL. Four and five parameter logistic calibration models are often used to fit the calibration curves for ligand binding assays.12 The curve fitting was performed by a five parameter regression model, plotting anti-PRP concentration versus A492nm. Triplicate curves included in each run permitted the evaluation of the intra-curve repeatability and the demonstration of the suitable intra-curve accuracy with %RE between 0.23 and 0.63%. The precision and accuracy inter-curves satisfied the acceptance criteria in the six assays, with CV≤9.78% and RE≤7.78%, respectively.
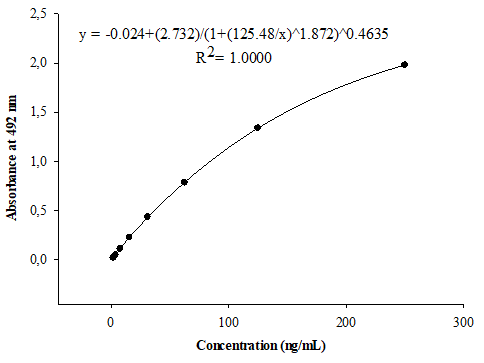
Figure 1 Calibration curve for the quantification of anti-PRP antibodies in serum, arranged with the complete dataset of six assays. Standard ranging from 1.95 to 250 ng/mL were prepared with the Human anti-Haemophilus influenzae b reference serum (NIBSC, UK).
Minimum required dilution
The MRD is the smallest dilution to which a sample must be diluted in the buffer to optimize accuracy and precision in an assay run, by reducing the signal to noise ratio.9 MRD typically ranges from 1:5 to 1:100.10 In this study, spiked samples were prepared in the same matrix as the study samples for determination of the MRD. The acceptance criteria for MRD was based on the percent of the recovery, determined by the relation between the back-calculated concentration for each point of the curve, prepared with a pool of ten serum samples spiked with NIBSC standard, and the nominal concentrations of the calibration curve. Using dilution factors upper than 20, the percentage of recovery was between 80 and 120%, and the best value of recovery was obtained with the dilution 1:80 (Table 1). However, the detectability of the assay at 1:80 was 0.156µg/mL of serum; this value is slightly higher than 0.15µg/mL, which is the minimum protective serum level of anti-PRP antibody.5 Thus, MRD should be lesser than 1:80. Data from Table 1 were used for the interpolation of 50 as dilution factor in the curve prepared with the spiked pool of sera and in the calibration curve. In 1:50 dilution point, the back-calculated concentration was 24.04 ng/mL and the nominal concentration, 25.0 ng/mL; with 96.17% of recovery. Attending to this result, 1:50 was selected as MRD, with a detectability of 0.120µg of anti-PRP antibodies per milliliter of serum.
Sample dilution factor |
Nominal concentration (ng/mL) |
Back-calculated concentration (ng/mL) |
Recovery (%) |
5 |
250 |
122 |
48.86 |
10 |
125 |
95.8 |
76.64 |
20 |
62.5 |
55.6 |
88.89 |
40 |
31.2 |
29.3 |
93.85 |
80 |
15.6 |
15.1 |
96.84 |
160 |
7.81 |
8.9 |
113.89 |
320 |
3.91 |
4.34 |
111.08 |
640 |
1.95 |
2.06 |
105.4 |
Table 1 Recovery analysis in the minimum required dilution assessment
Recovery (%)=(Back-calculated concentration/Nominal concentration) x 100
Specificity
High and low concentrations of NIBSC standard, diluted 1:50 in a matrix with low anti-PRP concentration, were incubated independently with an excess of synthetic antigen (PRP-HA) or natural antigen (HbOHA). Both antigens inhibited the reaction of soluble antibodies with the synthetic antigen coating the ELISA plate. The mean percent of inhibition was higher than 92% in all reactions, which supported the specificity of the assay to detect only the target analyte, in this case, anti-PRP antibodies, with the synthetic or the natural antigen. Some differences are marked when assessing specificity for ligand binding assays. Chapter of specificity is not given in FDA guidelines.10,12 Description related to specificity is contained in a chapter of selectivity briefly and not in detail. The distinctions between the agencies concern sample concentrations, type of matrix, and concentration of interfering molecules,13 but the acceptance criterion for the percent of inhibition has been not specified for regulatory guidelines. The use of subjective criteria, such as ≥50% inhibition of signal, is discouraged.14 We consider that a percent of inhibition higher than 90% is an acceptable criterion that supports the specificity of the immunoassay. To achieve this, the optimal conditions for the antigen-antibody reaction should be assurance during the inhibition step. The elimination of possible background noise could be one of the strategies to obtain a percent of inhibition close to 100%.
Selectivity
Selectivity is the ability of an analytical method to detect and differentiate the analyte in the presence of other components in the sample. In the present study, selectivity was assessed with hemolytic and lipemic sera, since the most common nonspecific interferences in the ELISA are due to hemolysis and lipemia.15 According to the EMA and FDA guidelines,9,10 at least ten individual matrices should be spiked at or near the lower limit of quantification to evaluate selectivity. In these experiments, ten individual hemolytic sera and ten individual lipemic sera were spiked with the NIBSC standard, to prepare pools with low concentrations of anti-PRP antibodies. The percent of the recovery, between the back-calculated concentration of the pool, and the sum of the anti-PRP concentration of the sample and the concentration of the spiked standard, was in the acceptance interval from 80 to 120% for both kinds of evaluated samples: from 87.45 to 118.10% for hemolytic and from 84.68 to 112.81% for lipemics. Thus, hemolysis and lipemia did not interfere in the ability of the ELISA to measure the anti-PRP antibodies in serum samples.
Accuracy and precision
The accuracy of an analytical method describes the closeness of the determined value obtained by the method to the nominal concentration of the analyte.9 In this study, accuracy was represented by the relative error expressed as a percentage, and it was determined with the back-calculated concentration, as the observed value, and the nominal value as the expected one. For the five spiked pools of sera, intra-assay and inter-assays accuracy were within the range of 0.61 to 18.1% and 2.55 to 10.7%, respectively, and both complied the acceptance criteria.
Intra- and inter-assay variability data were used to determine the precision of the ELISA. The values of CV (%) for intra-assay (3.21 to 7.06 %) and inter-assays (7.75 to 18.06%) fulfilled the acceptance criteria (Table 2). The precision of the assay was sufficient to allow the direct comparison of samples on the same and from different positions into the ELISA plate, at different days by two analysts. These results indicated that the method satisfied the specifications for accuracy and precision, following the guidelines for validation of immunoassays.9,10
Spiked pool of sera |
Mean back-calculated concentration (ng/mL) |
Variance Components |
CV (%) |
|
||
|
|
Analyst |
Day |
Error |
Intra-assay |
Inter-assays |
S1 |
1.9 |
0 |
0.055 |
0.018 |
7.06 |
18.06 |
S2 |
4.79 |
0.009 |
0.02 |
0.054 |
4.85 |
7.75 |
S3 |
22.7 |
0 |
3.328 |
0.53 |
3.21 |
8.65 |
S4 |
122 |
0 |
157.03 |
20.458 |
3.71 |
10.94 |
S5 |
223 |
6.062 |
225.77 |
70.977 |
3.77 |
7.79 |
Table 2 Results of the evaluation of intra- and inter-assays precision
CV (%), coefficient of variation expressed as percentage; CV intra-assay, (√variance of error) x 100/ mean of back-calculated concentration; CV inter-assays, (√∑variance components) x 100/ mean of back-calculated concentration
Quantification range
Because of the nonlinear nature of the standard curves in ligand binding assays, it is necessary to define both ends of the standard curve range to obtain the range of quantification, defined as the highest and lowest standard concentrations that meet the target precision and accuracy criteria.11 The anti-PRP concentration range was assayed from 0.0488 to 12.5µg/mL of serum. The intra-curve accuracy was between 0.07 and 12.96 %. The precision and accuracy inter-curves were generated by six repeated analysis of serially diluted standard. The lower point of the evaluated range, 0.0488µg/mL of serum, did not satisfy the acceptance criteria for accuracy and precision inter-curves (Table 3).
Nominal concentration |
Mean of back-calculated concentration (µg/mL of serum) |
RE |
CV |
(µg/mL of serum) |
from six assays |
(%) |
(%) |
12.5 |
11.4 |
-8.55 |
1.95 |
6.25 |
6.73 |
7.69 |
1.23 |
3.13 |
3.28 |
5.06 |
2.48 |
1.56 |
1.59 |
1.83 |
1.97 |
0.781 |
0.74 |
-5.25 |
3.16 |
0.391 |
0.401 |
2.54 |
5.14 |
0.195 |
0.183 |
-6.25 |
3.87 |
0.097 |
0.102 |
4.55 |
1.97 |
0.048 |
0.074 |
52.88 |
24.69 |
Table 3 Accuracy and precision inter-curves for the quantification range of anti-PRP concentration
%RE, percent of relative error; %CV, coefficient of variation expressed as percentage
The low limit of quantification was the lowest point for which the concentration was calculated on the regression curve with accuracy and precision below 20%. The upper limit of quantification was the highest point that met these criteria. Accordingly, the quantification range of the assay reached from 0.097 to 12.5µg/mL of serum.
Dilutional linearity
Dilutional linearity is assessed to demonstrate that a sample with a concentration above the upper limit of the quantification can be diluted to a concentration within the working range and still give a reliable result. At the same time, the presence of a hook effect is investigated.16 Demonstration of dilution linearity of samples containing the analyte of interest is a critical experiment to validate the specificity and accuracy of the method.17The curve for linearity assessment was performed with thirteen points, by two-fold serial dilutions of samples exceeding the upper limit of quantification. The best linear curve fit for the back-calculated concentration ranged from 2.07 to 213.46 ng/mL, with values of relative error <20% (Table 4), which rewarded the acceptance criterion.
Dilution points |
Dilution factor |
Nominal concentration (ng/mL) |
Back-calculated concentration (ng/mL) |
RE (%) |
Back-calculated concentration in serum (µg/mL) |
1 |
51 |
2000 |
533.51 |
-73.32 |
27.21 |
2 |
101.97 |
1000 |
456.74 |
-54.32 |
46.57 |
3 |
203.82 |
500.4 |
340.41 |
-31.97 |
69.38 |
4 |
407.14 |
250.5 |
213.46 |
-14.78 |
86.91 |
5 |
812.34 |
125.6 |
118.24 |
-5.85 |
96.05 |
6 |
1616.96 |
63.08 |
58.45 |
-7.33 |
94.51 |
7 |
3203.45 |
31.84 |
28.07 |
-11.84 |
89.92 |
8 |
6288.42 |
16.22 |
13.33 |
-17.81 |
83.82 |
9 |
12128.2 |
8.41 |
7.14 |
-15.1 |
86.6 |
10 |
22741.13 |
4.51 |
4.23 |
6.2 |
96.19 |
11 |
39960.24 |
2.55 |
2.07 |
18.82 |
82.72 |
12 |
64709.73 |
1.58 |
1.92 |
21.51 |
124.24 |
13 |
93738.37 |
1.09 |
1.97 |
80.73 |
184.66 |
Table 4 Results from experiments of dilutional linearity
RE (%), percent of relative error
Data from back calculated concentration in serum into the calibration range, revealed a mean of 89.59µg/mL of serum, with a standard deviation of 5.43µg/mL; the coefficient of variation was 6.06% and it satisfied the acceptance criterion for data precision. Hook effect was not observed in the assayed range, including the most concentrated sample, eight-fold higher than the maximum concentration point of the calibration curve. Consequently, the analyte of interest, in a concentration above the upper level of quantification, can be diluted to bring the analyte concentration into the calibration range.
Parallelism
Parallelism is a critical aspect to address relative accuracy by assessing the effects of dilution on the quantitation of analytes in a biologic matrix. The goal of investigating the parallelism is to ascertain that the binding characteristic of the endogenous analyte to the antibodies is the same as for the calibration analyte.18 Parallelism was assessed using seven pools of samples. This approach eliminated the generation of multiple values for individual study samples. Sample pool dilution curves were parallels to each other and the calibration curve (Figure 2). Data precision of the dilution series into the calibration range did not exceed 7.05 %, and it satisfied the acceptance criteria, as a confirmation of the comparable selectivity between the analyte from an endogenous sample and calibration analyte. Endogenous analyte concentrations provided the same degree of detection in the calibration curve after several dilutions. Therefore, the demonstration of parallelism indirectly confirmed the specificity of the ELISA. The specificity of the assay is considered uncertain if parallelism cannot be achieved within samples as well as between samples and calibration curve.18,19
Quality controls
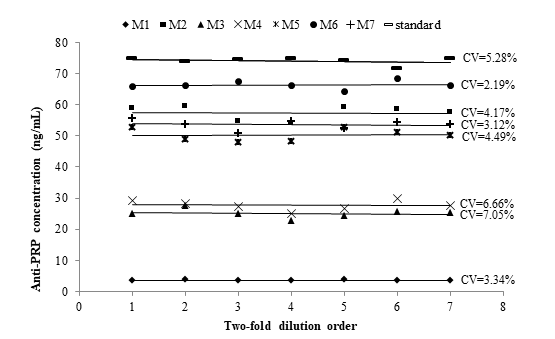
Figure 2 Parallelism assessment between the calibration curve (standard) and serially two-fold diluted pools of samples (M1 to M7) with high anti-PRP concentration. Pool dilutions were included into the calibration range. %CV: Coefficient of variation expressed as percentage.
The positive control must be generated from subjects’ samples, for use as QC during the routine performance of the immunoassay.10 The preferred QC samples are low, medium, and high patient pool samples. These are favored over spiked samples.20 In this study, the QCs were prepared with the patient pool samples with three anti-PRP concentration levels within the calibration range. The nominal anti-PRP concentration of each QC was determined using data from six assays performed by two analysts. All data fulfilled the precision criterion (Table 5). The low and high specification limits for QCs were established within±20% of their nominal concentrations (Table 6). The definition of these accuracy intervals will facilitate the use of this set of QCs in the clinical assays, to validate the acceptability of the results obtained over time.
Quality controls |
Mean conc. |
CV (%) |
Accuracy intervals |
||
(ng/mL) |
SD |
LSL (ng/mL) |
HSL (ng/mL) |
||
HQC |
97.8 |
6.3 |
6.44 |
78.2 |
117 |
MQC |
25.4 |
1.87 |
7.34 |
20.3 |
30.5 |
LQC |
8.39 |
0.89 |
10.62 |
6.71 |
10.1 |
Table 5 Inter-assays precision and accuracy intervals for quality controls
LQC, low-quality control; MQC, mid quality control; HQC, high quality control; Mean conc. (ng/mL), mean of anti-PRP concentration from six assays performed by two analysts; SD, Standard deviation; CV(%), coefficient of variation expressed as percentage; LSL, low specification limit; HSL, high specification limit
Quality Control |
Nominal conc. (ng/mL) |
6 days (2-8)ºC |
6h (22-25)ºC |
3 freeze-thaw cycles |
5 freeze-thaw cycles |
||||
Calc. conc. (ng/mL) |
RE (%) |
Calc. conc. (ng/mL) |
RE (%) |
Calc. conc. (ng/mL) |
RE (%) |
Calc. conc. (ng/mL) |
RE (%) |
||
HQC |
97.76 |
94.85 |
-2.98 |
107.6 |
1.05 |
98.97 |
1.24 |
102.8 |
5.15 |
100.1 |
2.42 |
109 |
11.49 |
95.64 |
-2.16 |
80.52 |
-17.6 |
||
95.07 |
-2.75 |
106.3 |
8.73 |
97.13 |
-0.64 |
93.14 |
-4.73 |
||
MQC |
25.44 |
27.19 |
6.87 |
25.18 |
-1.04 |
26.31 |
3.42 |
28.34 |
11.38 |
26.22 |
3.07 |
26.05 |
2.39 |
25.96 |
2.04 |
28.6 |
12.43 |
||
25.87 |
1.7 |
22.42 |
-11.8 |
24.92 |
-2.06 |
28.42 |
11.73 |
||
LQC |
8.39 |
8.14 |
-2.93 |
10.19 |
21.44 |
9.43 |
12.35 |
9.27 |
10.54 |
7.85 |
-6.49 |
9.88 |
17.8 |
9.58 |
14.17 |
9.88 |
17.8 |
||
|
|
10.19 |
21.44 |
8.22 |
-2.03 |
8.52 |
1.55 |
10.19 |
21.44 |
Table 6 Results of the evaluation of stability of quality controls at different temperatures and after freeze-thaw cycles
LQC, low quality control; MQC, mid quality control; HQC, high quality control; Calc. conc, back-calculated concentration; RE (%), percent of relative error
Robustness
The complexity of bioassays makes them particularly susceptible to variations in assay conditions, and it is essential to evaluate the robustness, assessed by the capacity of the assay to remain unaffected by small but deliberate variations in method parameters. For example, changes in temperature, incubation times, reagent lots, and buffer characteristics.9,10 The robustness of this ELISA was demonstrated after small variations in critical steps of the procedure. Changes in±0.2 mg/mL in the OPD concentration; the use of bovine serum albumin from four different suppliers for the preparation of blocking and dilution buffers; the use of Costar™ 3590 High Binding ELISA plate instead of Nunc™ MaxiSorp strip plate module; and the use of the lot 00064787 of HRP-conjugated goat antihuman IgG in place of lot 00073189, did not produce deviances from the QCs accuracy intervals (Figure 3). This was a confirmation of the capability of the ELISA to remain unaffected by small deliberate changes in method parameters, and it provided an indication of its reliability during normal run conditions.
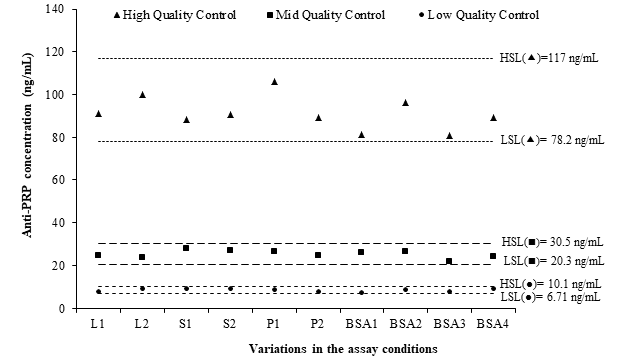
Figure 3 Evaluation of the robustness of the ELISA using the fulfilling of quality controls accuracy intervals.
LSL: low specification limit; HSL: high specification limit; L1: (HRP)-conjugated lot 00073189 (Dako, Denmark); L2: (HRP)-conjugated lot 00064787 (Dako, Denmark); S1: OPD 0.4 mg/mL; S2: OPD 0.8 mg/mL; P1: Costar™ 3590 High Binding plate; P2: Nunc™ MaxiSorp Strip plate module; BSA1: BSA AppliChem; BSA2: BSA Sigma; BSA3: BSA Spectrum; BSA4: BSA Roche.
Stability of quality controls and samples
Sample handling before, during, and after analysis has the potential to dramatically influence the results of a measurement. Therefore, it is important to investigate if different storage conditions contribute to systematic errors in order to provide the clinicians with adequate sample collection and transport instructions.16 In the present study, it was necessary to demonstrate the stability of QCs at the most used temperature conditions in clinical assays. If QCs have been prepared in the same matrix as the study samples, the use of QCs with high, mid, and low levels of antibody evaluates the stability of samples.10 The storage for 6 hours at (22-25)ºC and 6 days at (2-8)ºC, and three and five freeze/thaw cycles, did not affect the anti-PRP concentrations in QCs. QCs were considered stable because they fulfilled the accuracy intervals (Table 6). These results are valid for serum samples since they are from the same origin of QCs, which spans a range from low to high levels of anti-PRP antibodies.
An indirect ELISA for the quantification of antibodies against Haemophilus influenzae type b capsular polysaccharide was successfully validated according to regulatory guidelines. The results of the full validation show that the characteristics of ELISA fit well within the margins that define a reliable test. This ELISA represents a good tool fitting its purpose of analyzing the induced antibody response in children immunized with a conjugate vaccine with a PRP-synthetic antigen.
None.
Authors declare that there is no conflict of interest.
None.

©2021 Pérez-Bernal, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.