Journal of
eISSN: 2572-8466


Research Article Volume 5 Issue 2
1Department of Biochemistry, University of Delhi South Campus, India
2Department of Cell Biology and Molecular Genetics, University of Maryland, USA
3School of Biotechnology, Jawaharlal Nehru University, India
Correspondence: Suneel Kateriya, School of Biotechnology, Jawaharlal Nehru University, New Delhi, India
Received: December 26, 2017 | Published: March 26, 2018
Citation: Awasthi M, Ranjan P, Kateriya S. Cytoplasmic extensions of the channelrhodopsins 1 and 2 interacts in Chlamydomonas reinhardtii . J Appl Biotechnol Bioeng. 2018;5(2):84-90. DOI: 10.15406/jabb.2018.05.00121
Channelrhodopsin-1 (ChR1) and channelrhodopsin-2 (ChR2) are directly light-gated ion channels that are being extensively utilized in the field of optogenetics. These proteins are present in the eyespot of a phototactic alga called Chlamydomonas reinhardtii and mediate its motility behavior (phototaxis and photophobic responses). Both ChR1 and ChR2 are two seven transmembrane proteins with N-terminal rhodopsin domain and C-terminal cytoplasmic extensions of unidentified function. Earlier studies have suggested the interaction between ChR1 and ChR2. However, the evidences of physical interaction between ChRs are still lacking. Here, we have dissected the possibility of the direct interaction between the cytoplasmic C-terminus extensions of the ChR1 and ChR2. The immunoprecipitation, co-precipitation, co-localization and the proximity ligation assay showed the interaction between ChR1 and ChR2. In addition, the in vitro GST pull down assay showed that ChR1 and ChR2 interact directly with their C-terminal extensions. Our study leads to the model where C-terminal cytoplasmic extensions of ChR1 and ChR2 could directly interact with each other in Chlamydomonas. In vivo, this interaction could serve to directly regulate the activity or control binding to downstream effectors (Figure 1).
Keywords: channelrhodopsin, photoreceptors, chlamydomonas, protein‒protein interaction, proximity ligation assay
ChR1, channelrhodopsin-1; ChR2, channelrhodopsin-2; Cop, chlamyopsin; DIC, differential interference contrast; PLA, proximity ligation assay
Unicellular bi-flagellated green alga, Chlamydomonas reinhardtii exhibits two types of photomotility responses (phototaxis and photophobic responses).1,2 These two Photo behavioral responses help Chlamydomonas to obtain optimal light in the environment where these are present. Upon absorption of the photons by rhodopsins mediated Ca2+ flux appears in the eyespot region that bring about the primary electrical event called the photoreceptor current (PC), which depolarize the plasma membrane and serves as a trigger for the photomovement.1,3,4 The PC upon integration triggers the two flagellar current (FC) components called slow (Fs) and fast flagellar (Ff) currents that decides the flagellar beating pattern of microalgae.1,5,6
Photo behavioral responses in Chlamydomonas are mediated by two microbial type rhodopsin-based photoreceptors, which are composed of opsin (protein) and retinal that functions as chromophore.7,8 Out of twelve opsins-related genes identified so far in C. reinhardtii (named as Chlamyopsin1-12 or Cop1-12),9,10 Cop3 and Cop4 also named as channelrhodopsin-1/ChR111 and channelrhodopsin-2/ChR2.12 respectively, were found to be primarily responsible for the photo-behavioural responses of the Chlamydomonas reinhardtii.13‒15 Both ChR1 and ChR2 possess N-terminal seven transmembrane domain with light-gated cation channel activity16 and long C-terminal cytoplasmic extension of unknown function. ChRs regulate Ca2+ current in Chlamydomonas and thus the electrical process initiated by these photoreceptors regulates the photo sensory transduction in this microalgae.17,4 Electrophysiological measurements of ChR1 and ChR2 revealed the difference in the properties like wavelength sensitivity, desensitisation and on and off kinetics.18 At high light intensities ChR1 generates a photoreceptor current within <5μs indicating that it only involves the photo conversion of the photoreceptor protein. Whereas, at low light intensities another photoreceptor generates photocurrent within few milliseconds, which suggests the intermediate biochemical step between the receptor and channel activity.4,10
Immunoblotting and electrophysiological evidences revealed the up-regulation of ChR1 in the ChR2-RNAi cells, and that of ChR2 in the ChR1-RNAi strain, which indicated the crosstalk between ChR1 and ChR2 at genetic level.13 However, there is no direct evidence of interaction or cross-talk between ChR1 and ChR2 protein. Therefore, we hypothesize that ChR1 and ChR2 may interact directly or indirectly in C. reinhardtii cell and their interaction may serve as one of the critical intermediate step of the yet un-established downstream molecular mechanism that couples PC generation with flagellar movement during low light intensities.
In order to find out the interaction between ChR1 and ChR2, we utilized specific antibodies against the ChR1 and ChR2 proteins and established direct interaction between these two proteins via their C-terminus regions in the C. reinhardtii cell. In this report, we also established that the cellular distribution of ChR2 in Chlamydomonas is not confined to the eyespot only. That is, the cellular localization of ChR2 in C. reinhardtii is a light dependent process and it shuttles between the eyespot of Chlamydomonas to the basal bodies in light dependent manner. Moreover, like ChR1, which was earlier reported to be associated with the highly acetylated D4 rootlet in C. reinhardtii cells.19 In this study, ChR2 photoreceptor was also found to associate with D4 rootlet in C. reinhardtii cells.
Chlamydomonas strains and culture conditions
All the Chlamydomonas strains were obtained from the Chlamydomonas resource center (Department of Plant Biology, University of Minnesota). All the cell strains were grown in a the Tris-acetate-phosphate (TAP) culture medium under 14/10 h light-dark cycle in an incubator shaker at 24°C.
Antibody generation
Antibodies against C-terminus (Ct) of ChR1 and ChR2 were generated against the protein fragment from amino acid number 307-712 of ChR1 and 307-737 of ChR2, fused with 6XHis-Sumo tag (Sumo tag served as a solubility tag), respectively. ChR2-Ct and ChR1-Ct were purified under denaturing and non-denaturing conditions, respectively. Approximately 3 mg of purified proteins of ChR1 andChR2 was sent as an antigen to Genie Bangalore, India (Commercial facility for generating antibodies) for raising polyclonal antibodies in a rabbits.20 For generating peptide antibodies, peptide sequences of ChR1 and ChR2 were selected from the extracellular loop region of rhodopsin domain. Selected peptide sequences were the regions with minimum protein sequence identity between ChR1 and ChR2 as suggested by Multalin web-based server.21 The antigenicity of peptides was predicted by web-based Kolaskar & T ongaonkar antigenic prediction tool.22 The peptides were synthesized, and KLH (Keyhole Limpet Hemocyanin) was conjugated at C-terminus before injecting into the animal by Genie Bangalore, india. ChR2 specific peptide antibody was raised in mice whereas peptide antibody specific to ChR1 was raised in rabbit. Specificity of the each antibody was checked by immunoblotting using respective affinity purified full-length proteins expressed in E. coli.
GST pull-down assay
A nucleotide sequence corresponding to 307-737 a.a of ChR2 was cloned in frame with glutathione S-transferase (GST) in a pGEX-4T-1 vector (Invitrogen, Carlsbad, CA, USA). ChR2-Ct GST fusion protein was produced in E. coli, BL21 (DE3λ) cells by induction with 0.1 mM IPTG at 16°C for 48 h. For negative control, GST protein was affinity purified simultaneously under the similar conditions. ChR2-Ct-GST fusion protein and GST protein were affinity purified using glutathione sepharose 4B (HiTrap affinity columns, GE Healthcare, Little Chalfont, United Kingdom). Total cell extract of Chlamydomonas (CrTCL) was prepared by re-suspending 1g of freshly harvested Chlamydomonas culture pellet in 10 ml of cold IP-lysis buffer (20 mMTris-HCl pH8, 150 mM NaCl, 1% NP 40, 0.5% Sodium deoxycholate, 0.1% SDS). Lysate was sonicated by giving 8/10 s on/off pulses for 2 min and subsequently clarified by centrifugation at 13,300 rpm at 4°C for 15 min. Before GST pulldown, 1ml of lysate was pre-cleared by incubating it with GST using 20μl GST beads and 20μg of GST protein for 30 min at 4°C. Pull down assay, 1 ml of pre-cleared lysate was incubated with ChR2-Ct-GST protein and recombinant GST separately at 4°C in the rocking platform for 3 h. It was followed by incubation with GST beads for further 1 h. GST beads along with co-precipitated complex were separated by low-speed centrifugation at 3000 rpm for 5 min. Beads were washed five times using IP-lysis buffer and two times with PBS. The co-precipitated complex was eluted from the GST beads using elution buffer (50 mM Tris-Cl, 25 mM reduced glutathione). Loading samples were prepared in 2X Laemmli buffer by incubating at 60°C for 30 min. Protein was resolved with SDS-PAGE and analyzed by Immunoblotting.
Immunoblotting
Protein bands resolved in 10% SDS/PAGE were electroblotted onto the nitrocellulose membrane. Blocking of nitrocellulose membrane was performed with 5% non-fat dry milk powder in PBS supplemented with 0.1% tween-20 for 1 h at room temperature. Incubation with freshly diluted primary antibody (1:4000dilution), was carried out for 1 h at room temperature followed by incubation with HRP (horseradish peroxidase) conjugated anti-rabbit/ mouse secondary antibodies (1:5000 dilution).23 Immunolabelling was detected by enhanced chemiluminescence method, using the standard protocol.
Immuno precipitation
Peptide antibodies (~50µg) against ChR2/1 or pre-immune serum were mixed and incubated overnight at 4°C with pre-cleared Chlamydomonas total cell lysate in the presence of protease inhibitor cocktail. Protein A-sepharose beads were then split, mixed in the tubes containing total cell lysate along with specific peptide antibodies or pre-immune serum and incubated for 1 h at room temperature. Beads bound to the antibodies along with the protein complex were settled by centrifugation for 5 min at 3000 g. Beads with associated protein complex were washed three times with cold phosphate buffer saline (PBS). After complete removal of the supernatant, immuno precipitated proteins were eluted using 100µL of freshly prepared elution buffer (0.2% TFA). Elution fractions were incubated with 2 X Laemmli buffers (3% SDS) and incubated at 60°C for 30 min.
Immunolocalization
Chlamydomonas cells were harvested in early log phase by low-speed centrifugation (3000 g for 5 min). Cells were then gently re-suspended in PBS. Cell suspension aliquots of approximately 200µl (3 X 108 cells) were then seeded on acid washed cover-slips and fixed with 3.7% paraformaldehyde dissolved in PBS. Algal cells on the slides were permeabilized by submerging them in cold 100% ethanol and incubating cells adhered on cover-slips at -20°C for 10 min. Cells were then washed with PBS containing 0.25 M NaCl at room temperature for 10 min, followed by PBS for 5 min. After brief washings with 1 X PBS containing 0.5% Triton X-100 (PBST), incubation with freshly diluted primary antibody (1:125 dilutions in PBST), was carried out overnight at 4°C.17 Samples were then subjected to four successive washes in PBST and incubated with Alexa-Fluor 546–conjugated goat anti-rabbit IgG (Molecular Probes, 1:1000 dilutions in PBS) against primary antibodies raised in rabbit. Alexa-Fluor 488–conjugated goat anti-mouse IgG (Molecular Probes) was used as a secondary antibody against primary antibodies generated in mice. After incubation with secondary antibody at 37°C for 1 h samples were washed with PBST followed by brief washings with PBS, cover slips were mounted on slides applying anti-fade reagent (Slow Fade Gold, Molecular Probes). Slides were visualized by Leica TCS SP5 confocal microscope.
Proximity ligation assay (PLA)
PLA was performed using Duolink II Rabbit/Goat Kit (Sigma-Aldrich, St. Louis, Missouri, United States) according to manufacturer’s protocol with few modifications.24 C. reinhardtii cells were seeded in the glass cover-slip as mentioned inimmuno-localization procedure. After blocking, incubation with primary antibodies (ChR2-peptide and ChR1 peptide) was carried out overnight at ~4°C. PLA plus and minus probes (containing the secondary antibodies conjugated to oligonucleotides) were added and incubated at room temperature for 2 h. Further, ligase was added, and the ligation reaction was set at room temperature for 2 h. The ligation reaction was followed by incubation with an amplification-polymerase solution at room temperature for 2 h. After carrying out washings with the provided buffers, cover-slips were mounted on the glass slide in an inverted manner. Resulting red fluorescence signals were observed by the confocal microscope, Leica TCS SP5 (Leica Microsystems, Wetzlar, Germany) and visualized with excitation at 543 nm (emission, 585–615 nm).
ChR2 peptide antibody doesn’t cross-react with closely related ChR1 protein
In order to establish the interaction between ChR1 and ChR2, we generated ChR2 specific peptide antibody. We also utilized ChR1 specific peptide antibody that has been described and used in the earlier report by our group.25 With the help of the bioinformatics tools, 10a.a long peptide region of ChR2 was identified. This peptide region was found suitable for generation of antibody specific for ChR2 photoreceptor protein. The selected peptide region of ChR2 was from the extracellular loop region, present between the second and third helix of the rhodopsin domain. This unique peptide region possess no/least sequence identity with the sequence of ChR1 photoreceptor protein, and fitted the antigenicity criteria as per the Kolaskar & Tongaonkar antigenic prediction tool22 for the generation of specific antibodies. We named the peptide antibody against ChR2 as ChR2-peptide antibody and tested its specificity by immunoblotting of affinity purified full length ChR2 protein, expressed in E. coli. Since ChR1 and ChR2 shares more than 80% of protein sequence identity,26 the antibodies against the C-terminal region of ChRs (ChR1 and ChR2) cross-react with each other (Supplementary Figure 1A) (Supplementary Figure 1B) in several experimental conditions. Hence, to establish the specificity of ChR2-peptide antibody towards ChR2 protein only, we performed immunoblotting of full length ChR1 and ChR2 protein with ChR2-peptide antibody (Supplementary Figure 1C). Interestingly, ChR2-peptide antibody specifically identified the ChR2 but not the ChR1 protein. Thus, ChR2-peptide antibody was found suitable for performing ChR2 specific studies in the cell. It recognized ChR2 protein as a band of apparent molecular weight of 72 kD (Supplementary Figure 1D) in immunoblotting of C. reinhardtii total cell lysate and decorated the eyespot of C. reinhardtii in immunolocalization (Figure 2A). No signal was detected by immunoblotting (Data not shown) and cellular localization with anti-KLH which served as negative control (Figure 2B).

Figure 2 Cellular localization of ChR1 and ChR2 by respective peptide antibodies
A. Cellular localization of ChR1 and ChR2 using respective peptide antibodies. Red fluorescence at eyespot represents ChR1 and ChR2 in upper and lower panel, respectively
B. Immunolocalization with KLH as negative control
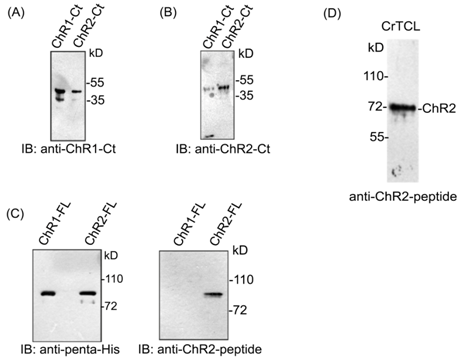
Supplementary Figure 1 Specificity of the anti-ChR2 peptide antibody.
ChR1 and ChR2 proteins interacts at the eyespot of Chlamydomonas
To find out the possibility of interaction between ChR1 and ChR2 proteins, we first performed the co-immunoprecipitation (co-IP) and then further validated the data using co-localization experiment. ChR1 was readily co-immunoprecipitated with ChR2 in IP using ChR2-peptide antibody (Figure 3A). Similarly, ChR1 was also co-immunoprecipitated with ChR2 in IP using ChR1-peptide antibody (Figure 3B). The presence of both the proteins co-immunoprecipitated with each other was validated by immunoblotting with ChR1 and ChR2 specific peptide antibodies, respectively. In order to further verify the possibility of interaction between ChR1 and ChR2, co-localization was performed. Although, we got some background signal in the co-localization experiment with ChR1 and ChR2-peptide antibodies but interestingly, ChR1 and ChR2 were identified to be predominantly present in the eyespot and were co-localized there with >87% Pearson correlation coefficient of co-localization (Figure 3C), upper and lower panels). These results indicated the possibility of direct or indirect interaction between ChR1 and ChR2 proteins. We therefore tested the possibility of direct or indirect interaction between ChRs using other methods of establishing the protein-protein interaction between two proteins.
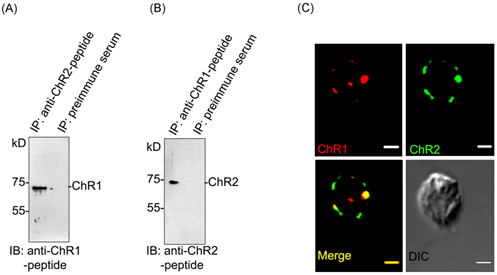
Figure 3Interaction of ChR1 with ChR2 in vivo.
ChR1 and ChR2 interacts via their C-terminus
In order to find whether the extended C-terminal (Ct) regions of ChR1 and ChR2 participate in interaction between ChR1 and ChR2, we performed the GST pulldown studies. C-terminal region of ChR1 (960-2136 bp) with N-terminal 6XHis-Sumo tag and ChR2 (945-2211 bp) with N-terminal GST tag were heterologously expressed in E. coli. Sumo-tag assists the solubility of ChR1 protein expressed in E. coli. The affinity purified ChR2-Ct-GST was used as a bait to pull down ChR1-Ct-6XHis-Sumo. Recombinant GST was used as bait in negative control experiment, performed under similar conditions. The presence of ChR1-Ct-6XHis-Sumo in the pull-down elutions was identified by immunoblotting with penta-His antibody. ChR1-Ct-Sumo was pulled down with ChR2-Ct-GST but not with recombinant GST (Figure 4A; upper panel). The presence of bait proteins (GST and ChR2-Ct-GST) in pulldown reaction were confirmed by immunoblotting with GST antibody (Figure 4A; lower panel). Thus pull-down assay showed that ChR2-Ct could directly interact with ChR1-Ct protein, in in-vitro conditions. To confirm whether the C-terminal region of ChR2 is sufficient to bind full length ChR1 protein, GST-pulldown of ChR1 protein from total cell lysate of C. reinhardtii was performed using ChR2-Ct-GST as bait and recombinant GST as negative control. Immunoblotting analysis of proteins bound to ChR2-Ct-GST and that to recombinant GST (negative control) with ChR1-peptide antibody showed the binding of ChR1 with ChR2-Ct-GST but not with recombinant GST protein (Figure 4B; upper panel). Immunoblotting with GST antibody confirmed the presence of bait proteins (GST and ChR2-Ct-GST), in respective lanes (Figure 4B; lower panel). The possibility of direct interaction between the ChR1 and ChR2 proteins was further confirmed by proximity ligation assay (PLA) using ChR1 and ChR2-peptide antibodies. The pre-immune serum of the animals (rabbit and mice, respectively) in which respective antibodies were generated, these were used as negative control for PLA, respectively. The presence of red fluorescence spot near the eyespot of C. reinhardtii cells suggested the possibility of the direct interaction between ChR1 and ChR2 (Figure 3C). The absence of any fluorescence signal in negative control experiment under similar experimental conditions served as negative control (Figure 3D). Thus, the proximity ligation assay of ChR1 and ChR2 strongly supported the interaction of these proteins in the eyespot of C.reinhardtii.

Figure 4 ChR1 and ChR2 interact via C-terminal extensions
Light affects the localization pattern of ChR2 in Chlamydomonas
In order to further explore the signaling cascade underlying the co-ordination between the photoreceptors that are present at eyespot and the flagellar beating, we mapped the cellular localization of ChR2 under different time points of 14/10 hr of light/dark cycle. After 14 h of light cycle, ChR2 was observed predominantly at the eyespot (Figure 5A; 14 h light). When cells were observed under dark cycle, we found that, in addition to the signal present in eyespot, ChR2-peptide antibody also decorated the basal bodies. Representative data has been shown after 1 h and 10 h of dark incubations (Figure 5A; 1 h and 10 h dark). However, after the light onset of 1h, we could not detect any signal of ChR2 at the basal bodies and it was again present in the eyespot only (Figure 5A; 1 h light). Quantification of the fluorescence intensity at the eyespot and basal bodies showed approximately 50% of increase in the ChR2 signal intensity in dark cycle (Figure 5B). The difference in the pattern of light dependent localization of ChR2 than that of ChR1,25 indirectly validated the ChR2 specific signal in C. reinhardtii. Thus light was identified to play magnificent role in the differential localization pattern of ChR2 in Chlamydomonas. To further explore the ChR2 localization in the C. reinhardtii cell and to find its relationship with highly acetylated D4 rootlet, immunolocalization via double labeling was performed. Chlamydomonas cells were immunostained with ChR2 peptide antibody and monoclonal anti-acetylated tubulin antibody at the same time. A large oval patch of ChR2 in eyespot associated with acetylated tubulin was observed in all the cells (Figure 5C). Thus, as observed for ChR1 in earlier reports, double labeling experiments showed the close association of ChR2 photoreceptor with highly acetylated D4 rootlet in C. reinhardtii cells.
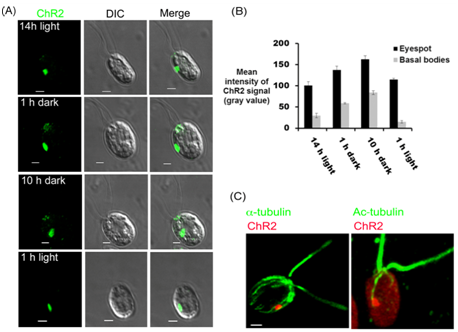
Figure 5 Light dependent cellular localization of ChR2 using the ChR2-peptide antibody.
Chlamydomonas channelrhodopsins (ChR1 and ChR2) are the two light gated ion channel proteins that functions as phototaxis receptor at high and low-light, respectively.13 ChR1 and ChR2 are highly homologous proteins that shares more than 80% protein sequence identity.11,26 Since, both possess N-terminal seven transmembrane region which are difficult to express in E. coli, the antibodies against the C-terminal (Ct) cytoplasmic tail has been extensively utilized in the field, for most of the in vivo and in vitro studies. We found that the antibodies generated against the C-terminal cytoplasmic tail of ChR1 and ChR2 shows cross reactivity and recognizes both the proteins in vitro. Therefore, in order to perform the ChR1 or ChR2 specific studies we generated specific antibodies against both the proteins that do not cross react with each other. The specific antibody against ChR1 has been used for ChR1 specific studies in the recent work by our group. However, in this study we utilized peptide antibodies against both antibodies to reveal probable cross-talk between both proteins, in-vivo. The photocurrent (PC) generation is a first step of the unidentified signaling cascade mediated by ChRs (ChR1 and ChR2). ChRs present at eyespot perceive the light (photons) and transfer of the photo-signals to the flagellar base. Suction pipette technique suggested that the PC generation comprises two different electrical events regulated by ChR1 and ChR2 which are named as early PC and late PC, respectively. At high light intensity early PC predominates that is generated within the micro-second time scale that suggests the direct interaction between rhodopsin and ion-channel. Whereas, at low light intensities late PC is generated in mili-seconds time scale that involves at least four orders of signal amplification and indicate the possibility of intermediate biochemical step(s) in signal transduction).10 The evidences of protein-protein interaction between ChR1 and ChR2, deciphered in this study suggested direct interaction between ChRs. It further supported the earlier evidences of interaction between ChR1 and ChR2, established by immunoblotting and electrophysiological measurements.13 The function of ChR1 and ChR2 in Chlamydomonas is well established, individually. The possible functional significance of the interacted identity of ChRs is yet to be determined. We hereby hypothesized the possibilities of the functional significance of ChRs interaction. ChR2, that triggers the phototaxis at low light intensity, might interact with ChR1 and this interaction might be a probable step to achieve the transmission of the optimal photosignal perceived at the eyespot of Chlamydomonas, which regulates flagellar beating, eventually leading to phototaxis. Broadly, the interaction of ChR2 with ChR1 might bring about changes in the optical and kinetic properties of these individual photoreceptors, turning them to be more adaptive to deal with the broad range of spectrum. Direct interaction of ChR1 and ChR2 could serve to regulate directly the activity of these photoreceptor proteins and might indirectly also control their association with downstream effectors.
Photocurrent is generated within the micro to milliseconds time scale of the light flash.10 Our result regarding ChR1 and ChR2 co-localization in Chlamydomonas cells, by fixing them with paraformaldehyde even after an hour of light exposure indicates the stable association between the two proteins. However, further investigation is needed to establish the nature of ChR1 and ChR2 association.
The cellular localization of the ChRs (photoreceptors) using antibody against ChR1 has been widely studied in Chlamydomonas.14,17,19,25,27 However, the cellular distribution of ChR2 specifically, remained sparsely appraised. We identified the involvement of light as an important abiotic factor that influences the localization of ChR2 in Chlamydomonas. ChR2 was present both in the eyespot and basal bodies of C. reinhardtii under dark cycle whereas it was mainly present in the eyespot during light cycle. Other than the effect of light in eyespot size and the light dependent expression and cellular distribution of ChR1, the differential distribution pattern of ChR2 might be the adaptive mechanism adopted by Chlamydomonas to respond to different light conditions where it is present. Literature suggests the association of ChR1 with the highly acetylated D4 tubulin rootlet which supports its microtubule based trafficking to the eyespot in Chlamydomonas. We found that ChR2 also associates closely with D4 rootlet in Chlamydomonas. Thus, this study further supports the model of the role of D4 rootlet in photoreceptor localization, established by the use of ChR1-Ct antibody.19,28 The mechanism of the trafficking of ChR2 from its site of synthesis to the eyespot and basal bodies might be characterized in the future study. However, identification of interaction between ChR1 and ChR2 has provided new insights into our understanding of the mechanism of the ChRs mediated photo signaling in Chlamydomonas.
Department of Biotechnology (DBT), Government of India is acknowledged for providing financial support of the projects (BT/PR9090/BRB/10/540/2007 and BT/PR10890/GBD/110/2008) to SK. MA and PR were supported with fellowship from University Grant Commission (UGC) Government of India. The CIF, UDSC facility is acknowledged for the technical support.
MA and PR carried out the experiments. SK supervised the experiments. MA and SK wrote the paper.

©2018 Awasthi, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.