Journal of
eISSN: 2572-8466


Research Article Volume 3 Issue 1
Bioinformatics Center, College of Fisheries, India
Correspondence: Roy AK, Bioinformatics Center, College of Fisheries,CAU (I) Lembucherra, Tripura, India
Received: January 27, 2017 | Published: April 21, 2017
Citation: Ghosh R, Upadhayay AD, Roy AK. In silico analysis, structure modeling and phosphorylation site prediction of vitellogenin protein from Gibelion catla . JAppl Biotechnol Bioeng. 2017;3(1):265-270. DOI: 10.15406/jabb.2017.03.00055
Vitellogenin is an egg yolk precursor protein expressed in the females of nearly all oviparous species including fish, amphibians, reptiles, birds, most invertebrates and monotremes .In the present study using bioinformatics tools and in silico modeling and analysis of Vitellogenin protein sequences of Gibelion catla was conducted. Primary structure prediction and physicochemical characterization were performed by computing theoretical isoelectric point (pI) 9.03, molecular weight 144862.03 Da, extinction co-efficient 78325, instability index 42.86 and aliphatic index 113.01. Grand Average of hydropathicity (GRAVY) was computed 0.212. Secondary structure assessment of Vitellogenin protein of carp Gibelion catla using GORIV reveals greater percentage of residues as alpha helix and random coils against the beta sheets. After performing homology modeling using Swiss model, a 3D structure of Vitellogenin of Gibelion catla have been predicted from its amino acid sequence. After the prediction structure has been validated through various validation tools. This homology modeling based structure will provide an insight to its functional aspects and further studies which are based on tertiary structure of protein.
Keywords: structure modeling, phosphorylation site, expasy peptide cutter, gibelion catla, vitellogenin, EDCs; APOB
Vitellogenin (VTG or less popularly known as VG) is a precursor protein of egg yolk normally in the blood or hemolymph only of females that is used as a biomarker in vertebrates of exposure to environmental estrogens which stimulate elevated levels in males as well as females.1 "Vitellogenin" is a synonymous term for the gene and the expressed protein. The protein product is classified as a glycolipoprotein, having properties of a sugar, fat and protein. It expressed in the females of nearly all oviparous species including fish, amphibians, reptiles, birds, most invertebrates and monotremes.2 Vitellogenin is the precursor of the lipoproteins and phosphoproteins that make up most of the protein content of yolk. In the presence of estrogenic endocrine disruptive chemicals (EDCs), male fish can express the Vg gene in a dose dependent manner. Vg gene expression in male fish can be used as a molecular marker of exposure to estrogenic EDCs. Vitellogenin precursors are multi-domain Apo lipoproteins (proteins that bind to lipids to form lipoproteins), that are cleaved into distinct yolk proteins.At the N-terminal, it contains a sequence of about 670 amino acids (LV1n chain), which (MTP) and apolipoprotein B-100 (APOB). It is usually synthesized by the extra ovarian tissues of female animals, secreted into bloodstream and transported to ovary, where it is internalized by growing oocytes and proteolytically cleaved to form yolk proteins that are later used as the nutrients by developing embryos and larvae3-8 However, Vg appears to have evolved pleiotropic functions in the advanced eusocial honeybee. It has been shown that honeybee Vg is associated with quite a few biological processes including social organization, temporal division of labor and foraging specialization, regulation of hormonal dynamics and change in gustatory responsiveness.9-12 Besides, honeybee Vg has been shown to be capable of reducing oxidative stress by scavenging free radicals, thereby prolonging lifespan in the facultatively sterile worker castes and reproductive queen castes.13 Another novel function of Vg is linked with immune defense.14 For example, Vg has recently been demonstrated to possess both hemagglutinating and antibacterial activities in the protochordate amphioxus (Branchiostoma belcheri) as well as the bony fish rosy barb (Puntius conchonius).15,16 SWISS-MODEL is a structural bioinformatics web-server dedicated to homology modeling of protein 3D structures. Homology modeling is currently the most accurate method to generate reliable 3-dimensional protein structure models and is routinely used in many practical applications. Homology (or comparative) modeling methods make use of experimental protein structures ("templates") to build models for evolutionary related proteins ("targets").6
The computed primary structure helps in quantitative study of protein-protein and protein-ligands interaction in solution. The aim to predict the secondary structure of protein based only on knowledge of their amino acid sequence. The secondary structure prediction displayed the alpha helix (H), beta strand (S), coil(C) and confidence score of prediction (0=low and 9=high for each residue). Structural class of protein can also be analyzed based on distribution of secondary structures elements. Long region of coil elements in the protein usually indicate unstructured disordered region. The interactions that occur between the C, O and NH groups on amino acids in a polypeptide chain to form α-helices, β-sheets, turns loops and other forms and that facilitate the folding into a 3-dimensional structure. 3D structure prediction By the Swiss model or other software is very easy within short time comparatively the output of experimentally determined protein structures typically by time-consuming and relatively expensive like X-ray crystallography or NMR spectroscopy is lagging far behind the output of protein sequences. The main aim of this study to analyze Vitellogenin protein and find utility of this protein in Gibelion catla.
Sequence analysis of vitellogenin protein of Gibelion catla (catla catla)
Vitellogenin protein of Gibelion catla was obtained from NCBI. The above obtained sequence was verified by Uniprot KB. The above obtained sequence was further used for complete protein sequence analysis (structural and functional annotation) and model building comparative modeling approach, Using ExPASy ProtParam Server complete primary structure analysis of protein has been performed, like molecular weight, theoretical isoelectric point (pI), amino acid composition, atomic composition, extinction co-efficient, half-life, aliphatic index, instability index and grand average of hydropathicity index (GRAVY). The ExPASy ProtParam tool is used to analyze protein sequences, structures and physicochemical properties of protein model.17
Secondary structure prediction
SOPMA and GORIV were used for secondary structure prediction of protein sequence of Vitellogenin.1 The comparative analysis of Vitellogenin protein by SOPMA and GORIV is given in the Table 1. And the result is graphically represented in Figure 1 & Figure 2.
Secondary Structure |
SOPMA |
GOR |
Alpha Helix |
51.76%(693) |
55.27%(740) |
310 Helix |
0.00% |
0.00% |
Pi Helix |
0.00% |
0.00% |
Beta Bridge |
0.00% |
0.00% |
Extended strand |
14.26%(191) |
9.78%(131) |
Beta Turn |
7.92%(106) |
0.00% |
Bend Region |
0.00% |
0.00% |
Random Coil |
26.06%(349) |
34.95%(468) |
Ambiguous States |
0.00% |
0.00% |
Other States |
0.00% |
0.00% |
Sequence Length |
1339 |
1339 |
Table 1 Percentage composition of all the secondary structure present in Vitellogenin of Gibelion catla.
Homology modeling of vitellogenin protein
The modeling of the 3-dimensional structure of the protein was done using by homology modeling programs3 Swiss model. Template was searched using Swiss model template library. A total 50 templates were found and it shows in Figure 3. The templates with highest quality have been selected for model building. Models are built based on the target template alignment model quality. Model quality was estimating assessing the QMEAN score.4 QMEAN is a composite scoring function for both the estimation of the global quality of the entire model as well as for the local per-residue analysis of different regions within a model.18 In the Swiss Model Workspace the QMEAN4 score is used to evaluate the generated models.
ExPASy peptide cutter
The main aim of this work is to design a protein which is easily digestible in human gastrointestinal tract and thus in silico digestibility was performed using the ExPASy Peptide cutter tool. Peptide cutter is used to predict the cleavage.19
Primary structure prediction and physiochemical characterization of protein
Using ProtParam tool provided by ExPASy primary structure have been performed it has been found that it is 1339 amino acid long residues of protein with 144862.03 Da molecular weight, whereas in Vitellogenin in Bactericera cockerelli (sulc) number of amino acid 2113 and molecular weight 234789.41 Da. In Gibelion catla theoretical isoelectric point or pI 9.03 which indicate its base property and Vitellogenin in Bactericera cockerelli (sulc) pI 9.18 which also base, whereas pI in Bovine α S1 Casein showed 5.77 which indicate that the protein is negatively charged and it can be precipitate in acidic medium. At 280 nm its extinction co-efficient has been calculated by the tool and it is found to be 78325 with its unit, on the other hand, the extinction co-efficient of Vitellogenin in Bactericera cockerelli (sulc) is 161650M-1cm-1 and the aquaporin protein from catfish is 36950M-1cm-1.The computed extinction co-efficient will provide help in quantitative study of protein-protein and protein-ligands interaction in solution.1 Another important parameter called instability index has also been calculated and is computed to be 42.86. And this classified the protein is unstable. In stability index provides the stability of protein in laboratory condition or in test tube. If the value of instability index is less than 40, then it is considered as stable protein,2 on the other hand Vitellogenin in Bactericera cockerelli (sulc) instability index shows 48.45 which also unstable, whereas aquaporin protein in catfish calculated instability index found 30.22 which indicate that the protein is stable and in Bovine α S1 Casein showed 56.03 which indicate also unstable. Aliphatic index of protein was found to be 113.01 which indicate this protein can survive at wide temperature range as the value is quite high, on the other hand in aquaporin protein showed 108.67 and Vitellogenin in Bactericidal cockerelli (sulc) 64.13. Grand Average of hydropathicity (GRAVY) was computed0.212. It provides interaction with water of a particular protein. Low value of GRAVY index indicates better interaction. Here it is 0.212 which indicates better interaction of Vitellogenin with water and Vitellogenin in Bactericera cockerelli (sulc) Gravy shows -0.904. On the other hand, 0.65 of aquaporin in catfish indicate better interaction with water. Estimated half-life of this protein is 30, whereas Vitellogenin of Bactericera cockerelli (sulc) also 30. The half-life of would be 30h in mammalian reticulocytes (in vitro), >20h in yeast (in vivo) and >10h in E. coli. The half life is the prediction of time taken by the protein to reduce to half of its amount in the cell after its synthesis in the cell. Total no. of atoms is 20896 and formula C6549H10704N1756O1848S39. Total number of negatively charged residues (Asp + Glu): 129, Total number of positively charged residues (Arg + Lys): 147. And maximum % of amino acid Alanine 14.7 % (197) and minimum % of amino acid tryptophan 0 .4% (6), whereas in Vitellogenin of Bactericera cockerelli (sulc) negatively and positively charged residues are 211 and 247.
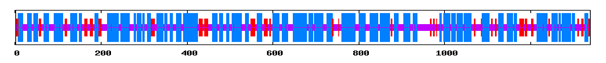
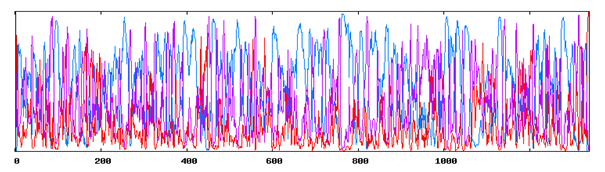
Figure 1 Secondary Structure Annotations of Vitellogenin from Gibelion catla using GORIV (h- Helix, e- extended strand, c-Random coil).
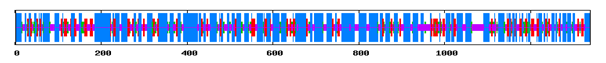
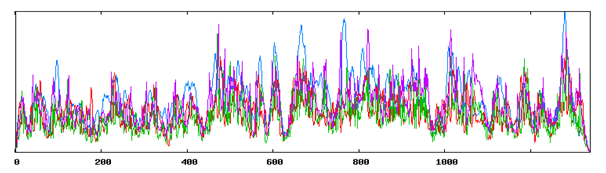
Figure 2 Secondary Structure Annotations of Vitellogenin from Gibelion catla using SOPMA (h- Helix, e- extended strand, c-Random coil).
Secondary structure prediction
Using SOPMA and GORIV (tool for secondary structure prediction) regions of different secondary structure have been located in the entire protein sequence and it was found that the secondary structure is composed of alpha helix and beta sheets. Table 1 represents the comparative analysis of GORIV and SOPMA from which it is clear that alpha helix is predominantly present when the structure is predicted by both SOPMA and GORIV, followed by extended strand. After secondary structural annotation analysis, it has been found that highest percentage is of alpha helix (51.76%) and then random coil (26.06%), extended strand has been found to be (14.26%), beta turn (7.92%). And other secondary structure like 310 helix, pi helix, Beta Bridge, bend region, ambiguous states and other states are totally absent. Higher percentage of helix indicates its helical chemistry (Figure 1 & 2) (Table 1).
Modeling of vitellogenin protein of Gibelion catla
3-dimensional structure of the protein (Figure 3) is predicted using the program called Swiss model using 1lsh.1 template from Swiss model template library for homology modeling. The evaluation of the predicted structure generated by Swiss model in Figure 4. Computational prediction of 3D structure of protein is based on Ab-initio, threading or Homology modeling. If the sequence similarity search of target sequence is more than 60%, then homology modeling can be useful for 3D structure prediction of target protein. Homology modeling was done using a template sequence whose structure has been solved by either X-ray diffraction or NMR Spectroscopy. Swiss model is online tool for 3D structure prediction based on homology modeling.4,20 This model quality is estimated by based on the Q mean score. Here Q means score Showed -3.70. 3D structure prediction By the Swiss model are very easy within short time comparatively the output of experimentally determined protein structures typically by time-consuming and relatively expensive like X-ray crystallography or NMR spectroscopy.21
ExPASy peptide cutter
For the proper metabolism, digestion and absorption of protein in humans, it has to be cleaved or hydrolyzed with digestive enzymes like Trypsin, pepsin, chymotrypsin, Thrombin, clostripain and lysine to yield peptides and free amino acids. The server shows the probability of the sequence cleaved by many enzymes. The total number of cleavages for chymotrypsin (high and low specificity), Arg-C protein’s, were found to be 68 and 261 Asp-N endopeptidase 129, Glutamyl endopeptidase 92, Clostripain60, Formic acid 37 which indicate the digestion rate of the protein is good.
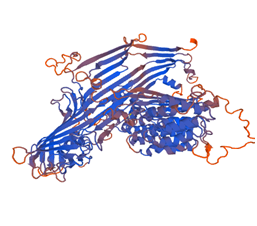
Figure 4 Model Structure of Vitellogenin protein of the Gibelion Catla 1lsh.1A (Lipovitellin as template).
Phosphorylation site prediction
Phosphorylation site has been located in the NetPhosK 3.1L Server. Different kinases like CAM-GSK3, CKII, cdc2, CK1, DNAPK, p38MAPK, PKG, ATM, RSK, PKA, cdk5, PKA, cdk5, PKB and UNSP have been found to be involved in the phosphorylation of protein. Highest score was predicted for the site 0.908 having serine residue. After analysis, % composition of amino acids has been obtained and is shown in Figure 5.
Scanprosite result
In Scanprosite result it showed that 1339 amino acid sequence are present and three disulphide bonds are present and their position between 260-290, 200 and 450 and it represent by Figure 6. Scanprosite is a new and improved version of the web-based tool for detecting PROSITE signature matches in protein sequences.22 Both user-defined sequences and sequences from the UniProt Knowledgebase can be matched against custom patterns or against PROSITE signatures.23 To predict protein function, assign family identity or detect remote homologues, searches against signature databases also known as secondary databases1 are essential, Scanprosite provides a web interface to identify protein matches against signature from the prosite database.2
Vitellogenin protein is one of the most important proteins, in this study Vitellogenin protein of Gibelion Catla was selected and it’s complete in silico analysis performed. Primary structure analysis and Physico chemical Characterization was performed by calculating various indices, GRAVY- 0.212 it is calculated by adding hydropathy values of each amino acid residues present in the protein sequence. The positive value indicated greater hydrophobicity and protein is sparingly soluble in water. The pI value can affect the solubility of a molecule at a given pH. Such molecules have minimum solubility in water or salt solutions at the pH that corresponds to their pI and often precipitate out of solution. Molecular weight 144826.03 Da and pI 9.03 which indicate that the protein is positively charged and that it can precipitate in base medium. Another important parameter called instability index has also been calculated and is computed to be 42.86. And this classifies that the protein Vitellogenin in Gibelion catla is unstable and easily able to degrade. Vg plays an integrative function in regulating immunity via its pleiotropic effects on both recognizing pathogen-associated molecular patterns and promoting macrophage phagocytosis. Our results suggest that fish Vg plays an integrative role in regulating innate immunity via its recognizing microbial cell wall constituents.
Authors are thankful to the Dean, college of fisheries, Central Agriculture University, Lembucherra, Agartala for encouragement and support. The Financial assistant by DBT, GOI, New Delhi, India for BIF project under which this study has been carried out, is duly acknowledged.
The author declares no conflict of interest.

©2017 Ghosh, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.