International Journal of
eISSN: 2573-2889


Research Article Volume 7 Issue 1
1Department of Genetics, Center for Biosciences, Federal University of Pernambuco, Brazil
2Center for Research in Biological Sciences - Nupeb, Federal University of Ouro Preto, Brazil
Correspondence: Maria Cidinaria Silva Alves, Department of Genetics, Center for Biosciences, Federal University of Pernambuco, Recife, PE, Brazil
Received: November 14, 2024 | Published: November 29, 2024
Citation: Alve MCS, Silva RCC. Genome-wide in silico analysis of alpha-hairpinin in tepary bean (Phaseolus acutifolius). Int J Mol Biol Open Access. 2024;7(1):158-162. DOI: 10.15406/ijmboa.2024.07.00188
This study characterized antimicrobial peptides of the α-hairpinin family in the genome of tepary bean (Phaseolus acutifolius). Ten sequences with the α-hairpinin motif (PaHrp1-10) were identified. These motifs were found in proteins with Kunitz_legume, Peptidase_C1/Inhibitor_I29, and Root_cap domains. Most PaHrps exhibited a signal peptide in the N-terminal region, except for PaHrp1, PaHrp5, and PaHrp10. The α-hairpinin motifs varied in length from 19 to 34 amino acids, with molecular weights ranging from 2.08 kDa to 3.74 kDa, and isoelectric points ranging from 3.58 to 9.13. In silico analyses showed that PaHrp7 has antifungal, antibacterial, and antiviral activities, while PaHrp1 and PaHrp9 exhibited antifungal and antibacterial activities. PaHrp6 demonstrated antifungal and antiviral activities, and PaHrp8 showed antibacterial and antiviral activities. PaHrp2 and PaHrp4 exhibited only antiviral activity; PaHrp5 and PaHrp10 showed antifungal activity; and PaHrp3 exhibited only antibacterial activity. The 10 PaHrp genes were mapped to six of the 11 chromosomes of the species, with Chr11 anchoring five genes (PaHrp5-9). The PaHrp motifs exhibited four conserved cysteine residues, forming two disulfide bridges characteristic of α-hairpinins. This study expands knowledge about antimicrobial peptides in P. acutifolius, suggesting their potential for therapeutic and biotechnological applications in agriculture and human health.
Keywords: In silico analysis, α-hairpinin famil, genomic, antimicrobial peptides
The α-hairpinin family of antimicrobial peptides was recently identified in plants, possessing a specific signature motif (C1XXXC2-X(n)-C3XXXC4).1–3 α-hairpinin proteins exhibit a secondary structure composed of two antiparallel α-helices connected by a loop (helix-loop-helix), stabilized in the tertiary structure by two disulfide bonds (Cys1-Cys4/Cys2-Cys3).4–6 α-hairpinin peptides are sources of bioactive molecules with a wide structural diversity and exhibit various biological activities.3 They demonstrate antifungal and antibacterial activity in several species, including barnyard grass (Echinochloa crus-galli),7 chickweed (Stellaria media),8 macadamia (Macadamia integrifolia),9 and wheat (Triticum kiharae).5 Among the α-hairpinin peptides with trypsin inhibitory activity are BWI-2a and BWI-2b from buckwheat (Fagopyrum esculentum),4 C2 from pumpkin (Cucurbita maxima)10 and VhTI from Veronica hederifolia.11 Additionally, some α-hairpinins possess ribosome-inactivating activity, such as Luffin P1 from sponge gourd (Luffa cylindrica).12 Due to these diverse biological activities, this family of short peptides, with potent antimicrobial activities and a relatively simple structure, is a promising candidate for biotechnological applications.2 Tepary bean (Phaseolus acutifolius) is a sister species of common bean (Phaseolus vulgaris L.), highly adapted to heat and drought.13–15 Native to the Sonoran Desert, this species possesses genetic characteristics that make it resilient to moderate thermal stress conditions, with a reduced genetic repertoire for disease resistance, consistent with its adaptation to arid and hot environments.16,17
The significant genetic similarity and shared content among Phaseolus species provide a valuable foundation for engineering climate adaptation in common beans, a critical crop for food security. 18,19 Additionally, tepary bean shows promise as an alternative protein source for regions facing hotter and drier conditions. However, its wider adoption remains limited due to challenges like insufficient marketing and less appealing culinary qualities.18 The objective of this study was to characterize the antimicrobial peptides of the α-hairpinin family present in the genome of tepary bean, analyzing their physicochemical characteristics, in silico biological activities, and three-dimensional structure.
Genome retrieval and α-hairpinin identification
The genome of P. acutifolius was obtained from Phytozome (ID: 580, NCBI taxonomy ID: 33129).20 The identification of sequences belonging to the α-hairpinin family was conducted using the Cysmotif Searcher21 with a cysteine motif pattern (C1XXXC2-X(n)-C3XXXC4) characteristic of this family.1,22
Sequence characterization
Candidate sequences were subjected to InterProScan (https://www.ebi.ac.uk/interpro/search/sequence/) to identify characteristic conserved domains of the α-hairpinin class. The presence of signal peptides was predicted using SignalP 6.0,23 while subcellular localization was predicted using the ProtComp 9.0 tool from Softberry Inc. (Mount Kisco, NY, USA). Subsequently, sequences were evaluated for molecular weight (MW), isoelectric point (pI), and grand average of hydropathicity (GRAVY) using ExPASy ProtParam software (http://web.expasy.org/protparam/). Conserved sequence patterns were analyzed through multiple sequence alignment using Clustal Omega.24
In silico biological activity analysis
Candidate sequences were evaluated for antimicrobial activity using CAMPR3, employing four different algorithms: Support Vector Machine (SVM), Random Forest (RF), Artificial Neural Network (ANN), and Discriminant Analysis (DA),25 in addition to ClassAMP.26 Antifungal activity was analyzed using Antifp27 and antiviral activity using Meta-iVP.28 Activity was considered significant for predictions with a score greater than 0.5 (50%).
After mining, 10 sequences containing the α-hairpinin motif from P. acutifolius (PaHrp1-10) were identified. This motif was found in proteins with Kunitz_legume (1), Peptidase_C1/Inhibitor_I29 (3), and Root_cap (6) domains (Table 1-S1), as observed in plant species such as the legume Peltophorum dubium.29 Most PaHrps exhibited a signal peptide in the N-terminal region, except for PaHrp1, PaHrp5, and PaHrp10. The α-hairpinin motifs varied in length from 19 (PaHrp7) to 34 (PaHrp2-4) amino acids (aa), with an average length of 31 aa. The molecular weight of the proteins ranged from 2.08 kDa (PaHrp10) to 3.74 kDa (PaHrp9), and the isoelectric point (pI) ranged from 3.58 (PaHrp4) to 9.13 (PaHrp7 and PaHrp9) (Table 1). α-hairpinins are low molecular weight antimicrobial peptides (10 kDa), and so far, few α-hairpinins have been reported, including MBP-1, MiAMP2s, Ec-AMP1, Luffin P1, VhT1, BWI-2c, Tk-AMP-Xs, and Sm-AMP-X.2,3 In lima bean (Phaseolus lunatus), PlHrp1 was present in 41 aminoacids, with a pI of 6.28 and molecular mass of 4.81.6 The 10 PaHrp genes were mapped onto the chromosomes of P. acutifolius (Figure 1). Out of the species' 11 chromosomes, only 6 contain PaHrp genes. Chromosome 11 (Chr11) harbors 5 genes (PaHrp5-9), while the remaining chromosomes each contain only one gene: Chr01 (PaHrp4), Chr04 (PaHrp1), Chr06 (PaHrp10), Chr07 (PaHrp2), and Chr09 (PaHrp3).
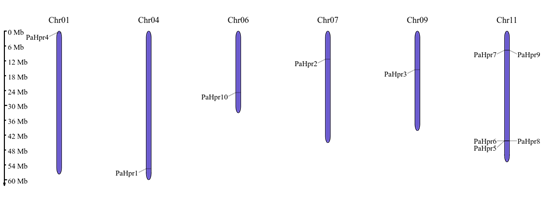
Figure 1 Distribution of PaHrp genes across chromosomes. The scale represents megabases (Mb). Chromosome numbers are indicated above each vertical bar.
|
|
|
Protein |
Motifs |
|||||
|
Gene |
Gene-ID |
Domain |
Signal peptide |
pI |
MW |
Length |
pI |
MW |
|
PaHrp1 |
Phacu.CVR.004G172300.1 |
Kunitz_legume |
Not |
8.85 |
23.01 |
31 |
4.86 |
3.48 |
|
PaHrp2 |
Phacu.CVR.007G109100.1 |
Peptidase_C1/ |
Yes |
8.95 |
50.54 |
34 |
3.66 |
3.69 |
|
Inhibitor_I29 |
||||||||
|
PaHrp3 |
Phacu.CVR.009G138200.1 |
Peptidase_C1/ |
Yes |
6.05 |
40.09 |
34 |
3.8 |
3.59 |
|
Inhibitor_I29 |
||||||||
|
PaHrp4 |
Phacu.CVR.001G009700.1 |
Peptidase_C1/ |
Yes |
6.16 |
39.75 |
34 |
3.58 |
3.63 |
|
Inhibitor_I29 |
||||||||
|
PaHrp5 |
Phacu.CVR.011G192600.1 |
Root_cap |
Not |
7.84 |
37.76 |
32 |
6.21 |
3.31 |
|
PaHrp6 |
Phacu.CVR.011G192300.1 |
Root_cap |
Yes |
8.24 |
40.61 |
33 |
6.17 |
3.5 |
|
PaHrp7 |
Phacu.CVR.011G088000.1 |
Root_cap |
Yes |
8.82 |
43.87 |
19 |
9.31 |
3.63 |
|
PaHrp8 |
Phacu.CVR.011G192500.1 |
Root_cap |
Yes |
7.84 |
37.76 |
32 |
6.21 |
3.59 |
|
PaHrp9 |
Phacu.CVR.011G088000.2 |
Root_cap |
Yes |
8.82 |
43.87 |
30 |
9.31 |
3.74 |
|
PaHrp10 |
Phacu.CVR.006G153300.1 |
Root_cap |
Not |
5.75 |
29.2 |
32 |
4.65 |
2.08 |
Table 1 Gene identification
aa, motif length; pI, isoelectric point; MW, molecular weight; GRAVY, average hydropathy index
|
Gene |
Sequence |
Size |
Pfam |
PaHrp1 |
MLHSETMKDSLLPFSILSFAFTIQLFIGIAVAAPEPVVDTSGQKLRTGVKYYILPVFRGKGGGLTVSSSGNNTCPLFVVQEKLEVLNGTPVTFTPYNAKSGVILTSTDLNIKSYGTTTSCDKPPVWKLLKVLTGVWFLSTGGVEGNPGIDTIVNWFKIEKAEKDYVISFCPSVCKCQTLCRELGLYVGDDGNKHLSLSDKVPSFRVMFKRA |
398 |
pfam00197 |
|
PaHrp2 |
MLNLTAACCHLSMRLQFTLLPLHYTLMYSPAPPFQGYNCNYIIHEENPIKRRRRRRRRKTKLVAEQAEEARTNMVAKRGHALSCFARISLVLFALTLSSARQTTVHDIAKKLKLEDNQLLRTEKKFNVFMENYGKKYSTREEYLQRLEIFAGNMLRAAENQALDPTAIHGVTQFSDLTEDEFQRHYTGVNGGFPWNNGVRDVAPPLKVDGLPEDFDWREKGAVTEVKMQGKCGSCWAFSTTGSIEGANFIATGKLLNLSEQQLVDCDNQCDITESTTCDNGCMGGLMTNAYKYLLQSGGLEEESSYPYTGAKGECKFDPGKVAVRITNFTNIPVDENQIAAYLVKHGPLAIGLNAIFMQTYIGGVSCPLICSKKWLNHGVLLVGYRAKGFSILRLGNKPYWIIKNSWGKRWGVDGYYKLCRGHGMCGMNTMVSAAMVTQTQTPSHNYASY |
398 |
smart00848 |
||||
|
PaHrp3 |
MATPSLFFLFSLLLFSATLVIANRIDGEDDLLIRQVVPDAEEHLLNAEHHFSAFKTKFGRTYATKEEHDYRFRIFKNNLLRAKSHQKLDPSAVHGVTKFSDLTPAEFSRQFLGLKPLRLPSDAQKAPILPTSDLPSDFDWRDHGAVTGVKNQGSCGSCWSFSAVGALEGAHFLSTGELVSLSEQQLVDCDHECDPEERGACDAGCGGGLMTNAFEYSLKAGGLMRENDYPYIGRDRGPCKFDKSRIAASVANFSVVSLDEEQIAANLVKNGPLAVGINAIFMQTYIGGVSCPYICGKHLDHGVLLVGYGAGAYAPIRFKEKPFWIIKNSWGESWGENGYYKICRGHNVCGVDSMVSTVAAIHTSSH |
263 |
smart00848 |
||||
|
PaHrp4 |
MARYTLCALLLFAAVAAAAAGASTDADDILIRQVVPEGEVEDHLLNAEHHFSTFKVKFGKTYATKEEHDHRFGVFKSNMRRARLHAQLDPSAVHGVTKFSDLTPAEFHRQFLGLKPLRLPAHAQKAPILPTNNLPKDFDWRDKGAVTNVKDQGSCGSCWSFSTTGALEGAHFLATGELVSLSEQQLVDCDHVCDPEEYGACDAGCNGGLMNNAFEYIIGSGGVQREKDYPYTGRDGTCKFDKSKIAASVSNYSVVSLDEEQIAANLVKNGPLAVAINAVYMQTYVGGVSCPYICGKHLDHGVLLVGYGEGAYAPIRFKEKPYWIIKNSWGENWGENGYYKICRGRNVCGVDSMVSTVGAIHASTQ |
341 |
smart00848 |
||||
|
PaHrp5 |
MEITKGSSIIIAVLLFVSCLSLQANAYYYRQCSTKGTRCYGKYIRCPNECPSSESTDPKAKVCQIDCDKPICRAVCRSRKPNCNAPGSGCYDPRFIGGDGRVFYFHGKTNEHFALVSDSSLQINARFIGHRPAGRARDYTWIQALGVLFNSKTLSLEAPKTSQWNEDVDHLKFTYNGNHLLLPQGPLSTWHSPQKDVKVERVAARNSVIVTLEDVAEILVNVVPVTKEDDAVHNYQVPQDDCFAHLEVQFRFFGLSPKVDGVLGRTYREDFENPAKVGVAMPVVGGEDKYRTTSLLSPNCASCVFSPPSSHHIEATEVSAELMGTLDCSKFSYGLGIVCKK |
341 |
pfam06830 |
||||
|
PaHrp6 |
MSFSINSDLLVFTSNWYLLVYKRKAMSVSSRNSLIYLLLLFAFCEMQIIAGKDTQTCISRKSPCFGKKVPCPDECPQKSPSDPKAKVCYLDCDSPICQAQCKTRKPNCNDRGSACLDPRFVGADGIVFYFHGRRNEHFALVSDVNLQINARFIGLRPATRTRDYTWIQALGLLFGSHKFTIEAIPAASWNDEVDHLKFSHNGKELAIPNGYLSTWQCPQNQLRIERTSSKNSVTITLPEVAEIFVNVVPVTNEDSRIHNYQIPKEDCFAHLEVQFKFHGLSSKVEGILGRTYQPDFQNPAKLGVAMAVVGGEDKYRTTSLVSADCGVCLFDGGKESSEKINSVSEYGLLDCTAAANSGNGIVCRR |
365 |
pfam06830 |
||||
|
PaHrp7 |
MGGKKWSILVAIFILLIAMEAAIAQGQGNGNDKGKGNENGNGNGKGKGSDNGKGKGEGSDDGKGKKNKKDDGKEKKPKEKKPKKQRDEASDYDKLSALPSGQERGFCRTNTTCEFKTIVCPSECAERKPKKNKKKKACFIDCSSSTCEATCKVRKANCDGYGSLCYDPRFVGGDGVMFYFHGAKGGNFAIVSDEEFQINAHFIGSRPQGRTRDYTWVQALGVMFDSHTLVIAANRVSHWNDKVDSLTVKWDGEVINVPTDGEAEWRANGDEREVVVERTDETNSVRVMVSGLVEMDISVKPIGEQENKVHNYQLPQNDAFAHLETQFRFKKSTDNFEGVLGQTYRPGYVSPVKRGVPMPMMGGENKYQTLSLFSTSCKRCMFQRPSSIASTEGLVAQY |
211 |
pfam06830 |
||||
|
PaHrp8 |
MEITKGSSIIIAVLLFVSCLSLQANAYYYRQCSTKGTRCYGKYIRCPNECPSSESTDPKAKVCQIDCDKPICRAVCRSRKPNCNAPGSGCYDPRFIGGDGRVFYFHGKTNEHFALVSDSSLQINARFIGHRPAGRARDYTWIQALGVLFNSKTLSLEAPKTSQWNEDVDHLKFTYNGNHLLLPQGPLSTWHSPQKDVKVERVAARNSVIVTLEDVAEILVNVVPVTKEDDAVHNYQVPQDDCFAHLEVQFRFFGLSPKVDGVLGRTYREDFENPAKVGVAMPVVGGEDKYRTTSLLSPNCASCVFSPPSSHHIEATEVSAELMGTLDCSKFSYGLGIVCKK |
450 |
pfam06830 |
||||
|
PaHrp9 |
MGGKKWSILVAIFILLIAMEAAIAQGQGNGNDKGKGNENGNGNGKGKGSDNGKGKGEGSDDGKGKKNKKDDGKEKKPKEKKPKKQRDEASDYDKLSALPSGQERGFCRTNTTCEFKTIVCPSECAERKPKKNKKKKACFIDCSSSTCEATCKVRKANCDGYGSLCYDPRFVGGDGVMFYFHGAKGGNFAIVSDEEFQINAHFIGSRPQGRTRDYTWVQALGVMFDSHTLVIAANRVSHWNDKVDSLTVKWDGEVINVPTDGEAEWRANGDEREVVVERTDETNSVRVMVSGLVEMDISVKPIGEQENKVHNYQLPQNDAFAHLETQFRFKKSTDNFEGVLGQTYRPGYVSPVKRGVPMPMMGGENKYQTLSLFSTSCKRCMFQRPSSIASTEGLVAQY |
366 |
pfam06830 |
||||
|
PaHrp10 |
MEHVCPNACPRGCEVDCITCKPVCKCDRPGAVCQDPRFIGGDGITFYFHGKKDRNFCLVSDPNLHINAHFIGRRNNNMKRDFTWVQSIAILFDNHQLFVGALRTPTWEDHIDRLALTFDGQPLTLYESEGATWTSSTVPNVSIVRTTSTNSVLVEVEGRLRVTAKVVPITEEDSRIHDYGITKEDCFAHLDLGFKFFTLSNEVSGVLGQTYKASYVSRVNVGANMPVMGGGKEFETTSLFSPDCSVARFIGKNELTEGDAFVS |
365 |
pfam06830 |
Table S1 Identification of complete sequences with the α-hairpinin motif in yellow, length, Pfam e domain.
Alignment results revealed that the PaHrp motifs exhibited four conserved cysteine (C) residues, forming two disulfide bonds with the characteristic pattern of α-hairpinins (Figure 2). α-hairpinins have a structure characterized by a helix-loop-helix motif, with four cysteine residues forming two disulfide bonds, thereby conferring stability to the structure.2,30 Despite showing high similarity in spatial structure, α-hairpinins exhibit low sequence homology and high functional diversity.3,30,31 These findings suggest that, despite the structural conservation of cysteines, variability in amino acid sequences may be related to the functional adaptation of these proteins, reflecting a wide range of specific biological activities.6 Analyses of in silico biological activities demonstrated that only PaHrp7 exhibited antifungal, antibacterial, and antiviral activities. PaHrp1 and PaHrp9 showed antifungal and antibacterial activities, PaHrp6 exhibited antifungal and antiviral activities, and PaHrp8 showed antibacterial and antiviral activities. PaHrp2 and PaHrp4 displayed only antiviral activity, while PaHrp5 and PaHrp10 showed antifungal activity, and PaHrp3 exhibited antibacterial activity (Table 2). In vitro antimicrobial assays demonstrated that maize α-hairpinin (MBP-1) contributed to grain resistance against infection caused by pathogenic fungi and bacteria such as Fusarium moniliforme and F. graminearum, as well as action against Escherichia coli and Clavibacter michiganense.32 Suppression of growth in filamentous fungi and bacteria was also observed in α-hairpinins from Stellaria media,8 Triticum kiharae,5 and Echinochloa crus-galli.33
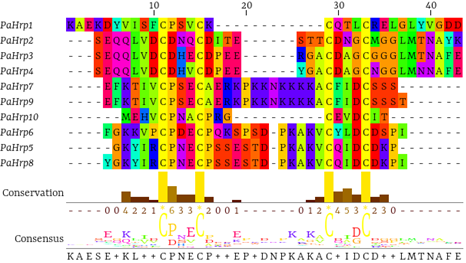
Figure 2 Multiple alignment of PaHrps demonstrating low sequence conservation, but revealing the conserved cysteines characteristic of α-hairpinins.
|
Gene |
ClassAMP |
ANTIFP |
Meta-iAVP |
|||||
|
SVM |
Score |
RF |
Score |
Prediction |
Score |
Prediction |
Score |
|
|
PaHrp1 |
Antifungal |
0.977 |
Antibacterial |
0.550 |
Antifungal |
0,200 |
Non-AVP |
0.406 |
|
PaHrp2 |
Antiviral |
0.778 |
Antiviral |
0.398 |
Antifungal |
0,039 |
AVP |
0.928 |
|
PaHrp3 |
Antibacterial |
0.711 |
Antibacterial |
0.408 |
Antifungal |
0,003 |
Non-AVP |
0 |
|
PaHrp4 |
Antiviral |
0.810 |
Antibacterial |
0.426 |
Antifungal |
0,115 |
AVP |
0.876 |
|
PaHrp5 |
Antifungal |
0.900 |
Antibacterial |
0.394 |
Antifungal |
0,285 |
Non-AVP |
0.294 |
|
PaHrp6 |
Antifungal |
0.939 |
Antibacterial |
0.484 |
Antifungal |
0,179 |
AVP |
0.524 |
|
PaHrp7 |
Antifungal |
0.905 |
Antibacterial |
0.612 |
Antifungal |
0,635 |
AVP |
0.962 |
|
PaHrp8 |
Antibacterial |
0.940 |
Antibacterial |
0.436 |
Non-Antifungal |
-0,095 |
AVP |
0.630 |
|
PaHrp9 |
Antifungal |
0.894 |
Antibacterial |
0.610 |
Antifungal |
0,713 |
Non-AVP |
0.316 |
|
PaHrp10 |
Antifungal |
0.934 |
Antibacterial |
0.454 |
Antifungal |
0,641 |
Non-AVP |
0.016 |
Table 2 Antimicrobial and antiviral potential of PaHrps.
SVM, support vector machine; RFC, random forest classifier, DAC, discriminant analysis classifier
This study significantly expands the understanding of antimicrobial peptides in P. acutifolius, particularly by characterizing α-hairpinin motifs across 10 identified PaHrp sequences. The structural versatility, despite low sequence homology, suggests an adaptive functional role that could be harnessed in various applications. The unique distribution of PaHrp genes across multiple chromosomes and the ability of certain peptides, like PaHrp7, to exhibit multi-target antimicrobial activity further underscore the potential of P. acutifolius peptides as valuable resources for developing therapeutic agents and enhancing crop protection strategies. These findings support future research into the biotechnological applications of PaHrps in agriculture and human health, paving the way for new avenues in natural antimicrobial development.
We thank ChatGPT for assistance with the translation of this article. We also thank CAPES (Level Personnel Coordination Superior, Brazil) and CNPq (National Council for Scientific and Technological Development) for fellowships and financial support.
The authors declared that there are no conflicts of interest.

©2024 Alve, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.