International Journal of
eISSN: 2573-2889


Research Article Volume 3 Issue 3
Donetsk Physical and Technical Institute, Ukraine
Correspondence: Helen A Grebneva, Donetsk Physical and Technical Institute, NAS of Ukraine, 03680 Kiev, Ukraine
Received: April 18, 2018 | Published: May 30, 2018
Citation: Grebneva HA. A polymerase-tautomeric model for radiation-induced genomic instability: targeted delayed base substitution mutations during errorprone and SOS replication of double-stranded DNA, containing cis-syn cyclobutane cytosine dimmers. Int J Mol Biol Open Access. 2018;3(3):125-141. DOI: 10.15406/ijmboa.2018.03.00065
Ionizing radiation and ultraviolet light cause genomic instability, which is transmitted through many generations after the exposure through the offspring of surviving cells. There are no clear mechanisms of genomic instability and delayed mutations formation I suggested, and develop alternative, the polymerase-tautomeric model of ultraviolet mutagenesis. A polymerase-tautomeric model for mechanism of targeted delayed base substitution mutations caused by cis-syn cyclobutane cytosine dimers is proposed. Ultraviolet radiation can lead to a change in the tautomeric state of DNA bases. Thus, cytosine may form seven rare tautomeric forms, which will be stable if the corresponding nucleotides are part of cyclobutane dimers. Structural analysis of the insertion of the bases showed that opposite rare tautomeric form of cytosine C4* can be incorporated, but may be inserted any other canonical base so that between them hydrogen bonds are formed. If in the synthesis of DNA containing the cis-syn cyclobutane cytosine dimmer CC4*, involved DNA polymerases with relatively high fidelity of synthesis, mutations not appear. However, if further DNA synthesis will involve DNA polymerases having a low fidelity of synthesis, there may be base substitution mutations. Moreover, they may be formed through many cycles of replication after DNA has been damaged. Consequently, these are the delayed mutations. Canonical cis-syn cyclobutane cytosine dimers CC may result in targeted delayed trans versions C-G®T-A only, cis-syn cyclobutane cytosine dimers CC4* may result in targeted delayed transitions C-G®T-A, targeted delayed trans versions C-G®A-T and C-G®G-C.
Keywords: radiation-induced genomic instability, rare tautomeric forms of DNA bases, cis-syn cyclobutane cytosine dimers, delayed base substitution mutations, error-prone replication, SOS replication
The conventional view of radiation mutagenesis is that radiation induces most mutations in cells shortly after irradiation.1 Radiation, including ionizing radiation such X-rays and charged particles (heavy ion radiation),1 as well as no ionizing radiation (UV light)1–3 and the DNA alkylation agent ethyl methane sulphonate,2 chemotherapeutic drugs,4 and photodynamic treatment5,6 induce genome instability many cell generations after the exposure. These delayed effects are observed after high (1-10Gy) and very low (0.01-0.1Gy) doses of ionizing radiation.1 Experimental data suggest a specific, and perhaps unique, role for radiation-induced genome instability as a critical early event associated with initiation of the carcinogenic process. In other words, radiation-induced genome instability is a critical early event in the multi-step sequence leading to radiation-induced cancer.7 Radiation-induced genome instability and radiation-induced bystander effects have been described in reference.8,9 Delayed mutations are mutations occur in the progeny of the irradiated cell after many generations of cell division.8 Delayed effects include hyper mutation, hyper-homologous recombination, chromosome instability and reduced clonogenic survival (delayed death).1 Thus, according to the definition,8 the radiation-induced genome instability in a narrow sense includes only delayed mutations. These include both targeted delayed mutations, as well as untargeted delayed mutations. Delayed mutations and untargeted mutations are two features of genomic instability.10 The genome instability is observed at the nucleotide level and results in base substitutions or deletions or insertions of a few nucleotides. In most cancers, the genome instability is observed at the nucleotide and chromosome level.11 Radiation-induced genome instability in the narrow sense includes targeted and untargeted point delayed mutations.8 In this article, the radiation-induced genome instability will be considered in the narrow sense of the word. In order to understand how targeted delayed base substitution mutations can be formed, we will study which mutations can appear opposite cis-syn cyclobutane pyrimidine dimers. Cis-syn cyclobutane pyrimidine dimers are DNA lesions that most often appear when the DNA molecule is irradiated with ultraviolet light.12 Ultraviolet mutagenesis is fairly well studied, ultraviolet light leads to a small amount of photodamage, in addition, it causes skin cancer. Therefore, it is an excellent model for studying the nature and mechanisms of any types of mutations formation that appear under the action of any mutagens.
Radiation-induced genomic instability under ultraviolet light
Ultraviolet irradiation of cells can induce a state of genomic instability that can persist for several cell generations after irradiation. However, questions regarding the time course of formation, relative abundance for different types of ultraviolet radiation, and mechanism of induction of delayed mutations remain to be answered.13 At present, the molecular mechanisms underlying the radiation-induced genome instability are not known.8 UVR comprises three different bands; long-wave UVA (320-400 nm), middle-wave UVB (290-320 nm), and short-wave UVC (200-290 nm).14 About 95% of solar radiation, which is the main source of UVR in environment is UVA and about 5% is UVB. UVC is almost entirely absorbed in the upper part of the stratosphere .14 Dahle & Kvam15 have shown that delayed mutations arise many cell generations after three types of carcinogenic irradiation: (a) UVA-; (b) UVB-; or (c) X-radiation. The frequency of mutations was measured 14 days after irradiation. The frequency of mutations was 10-7500-fold above background. The proportion of unstable clones was higher for the cells treated with UVA (13.2%) than for cells treated with UVB (9.2%) and X-radiation (9.6%). UVA-radiation, which is suspected to cause melanomas, produces few immediate mutations but more delayed mutations than UVB or X-radiation. The oxidative UVA-radiation did not.15
Ultraviolet light induces delayed mutations that appear at the fourth, sixth, seventh, eighth, ninth, and eleventh cell division after UV light exposure.16 For UVA radiation the deletion spectra were similar for delayed and early mutations.16 Delayed genome instability of cells exposed to UV radiation was investigated in reference.17 All mutations were small (1- to 2-base) changes, including substitutions, deletions, and insertions, and the majority were compound, with an average of four mutations per mutant. Large deletions were absent.17 The classical paradigm of radiobiology is based on the concept that all the effects of radiation on living matter are due to the direct action of radiation. It is believed that radiation-induced genome instability, including targeted and untargeted delayed mutations18–21 simply cannot be explained on the basis of direct DNA damage.22 According Averbeck the discovery of untargeted and delayed radiation effects has challenged the classical paradigm of radiobiology.23 Delayed effects can play an important role in the process of radiation carcinogenesis.10 Watanabe assume that a radiation cancer-causing target is protein. The hypothesis that signaling mechanisms play an important role in genomic instability.24 Thus, at present the mechanism of delayed mutations formation is not clear.8,18–21
Some features of the ultraviolet mutagenesis
Ultraviolet (UV) radiation produces cyclobutane pyrimidine dimers and pyrimidine-pyrimidone (6-4) photoproduct (from 78% to 84%), hydrated bases (1%-7%) (cytosine bases form more often), linked DNA-protein and DNA-DNA, and break in filaments (less than 1%) in Escherichia coil, yeast, in xeroderma pigmentosum variant cell extracts and in mammalian cells under induced by UVB irradiation.25,26 Cis-syn cyclobutane pyrimidine dimers account for a large majority of UV-induced mutations.11 In addition to pyrimidine-pyrimidine adducts, the purine-pyrimidine adducts are formed, for example, in consecution TpA, but they form very rarely.27 In cells of E. coli, irradiation with a wavelength of 260 nm produces about 40% of thymine dimers, 5%-19% of cytosine dimers and 19%-22% of dimers consisting of cytosine and thymine. For UVC and UVB, the total relative proportion of cyclobutane pyrimidine dimers formed in the TT, TC, CT and CC sites was about 28%, 26%, 16% and 30%, respectively.28 However, for UVA, cyclobutane pyrimidine dimers were formed much more frequently in the TT sites than at the TC, CT, or CC sites (57% vs. 18,11 and 14%, respectively).28 Cyclobutane pyrimidine dimers are effectively removed by excision repair.29 If not all dimmers are moved cyclobutane pyrimidine dimers and pyrimidine-pyrimidone (6-4) photoproduct may produce mutations.30 Mutations occur during error-prone and SOS replication, repair or transcription.31–34 They cause targeted base substitution mutations,35,36 targeted insertions,37 targeted deletions,37 targeted complex mutations38 and targeted delayed mutations.8,23,39,40 Only 5-12 % of cyclobutane dimers and (6-4) adducts result in replication errors.41 Usually mutations occur opposite the photoproducts.31–34,41 Such mutagenesis is termed targeted.41,42 Sometimes mutations are formed in the vicinity of the damage, a process that is termed untargeted mutagenesis.43 Sometimes delayed mutations are formed.8,23,39,40 Long-wave ultraviolet UVA light can cause delayed mutations.2,19 The delayed mutations are usually point mutations, more than half of them are base substitution mutations.44 As the experiment shows, DNA damage leading to delayed mutations is usually not removed. Delayed mutations can make a significant contribution to genetic diseases.45
Models of mutagenesis
At present, the conventional paradigm relates the reason of mutations exclusively to sporadic errors of DNA polymerases. It is assumed that the mutations arise because the DNA-polymerase sometimes incorporates non complementary nucleotides opposite the cyclobutane pyrimidine dimers.46–48 One hypothesis is that UV-induced mutations occur only after domination of the cytosine or 5-methylcytosine within the pyrimidine dimer.49–51 Oxidative damage of the bases is considered the cause of mutagenesis under the action of long-wave ultraviolet light (UVA).48 According to reference52 7,8-dihydro-8-oxoguanine makes an important contribution to the genotoxicity of UVA irradiation of the yeast Saccharomyces cerevisiae. In the paper by Watson & Crick53 it was stated that the spontaneous mutagenesis is based on capability of nucleotide bases to change their tautomeric state, which influences the character of base pairing. The participation of rare tautomeric forms in mutagenesis was repeatedly discussed.54–57 A large number of works devoted to the study of rare tautomeric forms have been performed, both in DNA bases and in model molecules.58 It was shown that after cytosine was irradiated with UV light (cytosine was isolated in a low-temperature argon matrix), it changed from the main tautomeric form to rare tautomeric forms, their ratio depended on the intensity of irradiation.59 In reference58 the nature of the defect states in crystals of bases of nucleic acids irradiated with UV light by the method of thermally stimulated luminescence was studied. It was concluded that there are rare tautomeric forms of cytosine in the investigated crystals.58
However, all currently existing mutagenesis models cannot explain most of the phenomena of mutagenesis.60,61 I have proposed and attempted to construct polymerase-tautomeric models for UV-induced mutagenesis60–80 radiation-induced bystander effects62,66,73,74 and radiation-induced genomic instability,72,75 based on idea by Watson & Crick53 that changes in tautomeric state are possible for DNA bases. A mechanism for changes in the tautomeric state of base pairs has been proposed ДНК for the case when DNA is UV-irradiated63 and cyclobutane pyrimidine dimers are formed.63 Five new rare tautomeric conformations of A:T63 and seven new rare tautomeric conformations of G:C60 base pairs are proposed that are capable of influencing the character of base pairing. The rare tautomeric forms of bases are stable when the respective bases are involved in cyclobutane thymine dimers and in DNA synthesis.64 The rare tautomeric forms of bases are stable if the corresponding bases are located near cyclobutane pyrimidine dimers (2-3 bases).62,66,73 I propose mechanisms of targeted base substitution mutations61,64,73 targeted insertions,61,68 targeted deletions61,69 and targeted complex insertions61,70 formation during error-prone or SOS synthesis of DNA containing cis-syn cyclobutane thymine dimers. I propose mechanisms of targeted base substitution mutations60 and targeted insertions,67 Formation during error-prone or SOS synthesis of DNA containing cis-syn cyclobutane cytosine dimers.
Cis-syn cyclobutane thymine dimers TT1*, TT4* и TT5* wherein a thymine occurs in the rare tautomeric forms T1*, T4*, or T5* were shown to cause targeted base substitution mutations only.61,64 Cis-syn cyclobutane thymine dimers TT2* wherein a thymine is in the rare tautomeric form T2* may result in targeted frames hift mutations (targeted insertions 61,70 and targeted deletions).61,69 A DNA site containing cis-syn cyclobutane thymine dimers TT2* and TT1*, TT4*, and TT5* may result in targeted complex frames hift mutations61,70 Cis-syn cyclobutane thymine dimers TT3* wherein a thymine is in the rare tautomeric form T3* may result in targeted delayed base substitution mutations72,75 Polymerase-tautomeric model for radiation-induced bystander effects - untargeted substitution mutations formation also has been developed.62,66,74 A polymerase-tautomeric model for radiation-induced genomic instability-targeted delayed base substitution mutations72,75 has been developed. A polymerase-tautomeric model for ultraviolet mutagenesis was proposed for the formation of hot and cold spots of UV-induced mutagenesis.65 The idea of Watson & Crick77–79 was tested. It is observed a dGTP/dT77 and a dCTP/dA mispairs78 with one of the bases in rare tautomeric forms in the active site of DNA polymerases. These structures unambiguously demonstrate that DNA bases in rare tautomeric forms can exist in the polymerase active site, providing strong support of the rare tautomer hypothesis and polymerase-tautomeric models of the authors through direct structural evidence.77–79
Analysis of mutagenesis models
Polymerase paradigm,46–48 tautomer model by Watson & Crick53–56 and deamination model49–51 claim an explanation targeted base substitution mutations only. The idea of Watson & Crick53 that rare tautomeric forms of DNA bases can play an important role in mutagenesis53 is certainly magnificent, but requires further development. The mutagenesis models resting on rare tautomer hypothesis are, in fact, physical-chemical. Usually, they are simply stating that a change in the tautomeric state of bases is possible and concluding an erroneous pairing and, consequently, base substitution mutation to result from that change. The biological data are, as a rule, ignored. Spontaneous mutagenesis models suppose that the tautomeric state of DNA bases can be
In the polymerase paradigm, it is assumed that both matrix and inserted bases are in canonical tautomeric forms. We make a structural analysis of the inserting of the canonical bases found in the study of A-rule in the papers83–87 opposite cis-syn cyclobutane (Figure 1A) thymine dimmers or (6-4) adducts. The canonical tautomer of adenine (Figure 1B) and the canonical tautomer of cytosine (Figure 1C) cannot form hydrogen bonds with canonical tautomers of cytosine for steric reasons. But canonical tautomeric forms of thymine can be incorporated opposite the canonical tautomer of thymine (Figure 1D). Specialized and modified DNA polymerases incorporates canonical bases capable of forming hydrogen bonds with demonized bases in template DNA.64 Therefore, the polymerase paradigm cannot explain the mechanism for the formation of targeted base substitution mutations. To understand the mechanisms of the formation of different mutations under the action of different mutagens, it is necessary to understand what happens when at least one mutagen acts. As such a model, in my opinion, ultraviolet mutagenesis is best suited. I believe that in order to understand how mutations are formed under the action of ultraviolet light, the following should be done. It is necessary to study the processes that occur when an ultraviolet quantum of energy interacts with a DNA molecule. It is necessary to see to what chemical changes of DNA structure this can lead. It is necessary to study the conditions under which these chemical changes will be stable. It is necessary to study what mutations they can lead to in the case of error prone or SOS replication of DNA that has such damage. This plan was implemented in several article cycles.60–80,88–92
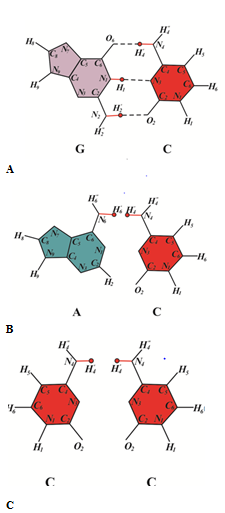
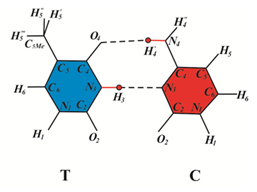
Figure 1 A structural analysis of pairing of canonical tautomer of cytosine C with canonical tautomer of DNA bases: (A) with guanine; (B) with thymine; (C) with guanine; (D) with cytosine.
A semi empirical potential function capable of describing hydrogen bonds with lengths different from equilibrium has been developed.88 It was used to find potential curves of the guanine-cytosine pair for several lengths of hydrogen bonds.89,90 The obtained curves were used to study the nature of the vibrations of atoms and atomic groups for isolated bases and guanine-cytosine base pair for hydrogen bonds in the ground and excited states.89,90 The problem was solved for an isolated guanine-cytosine pair89 and a pair of guanine-cytosine located in the DNA strand.90 The theory of heat de excitation of hydrogen bond protons in paired bases of DNA molecules was developed.91 The previously obtained results88–90 made it possible to estimate the lifetime of the excited hydrogen bond with respect to thermal transitions.91 The processes of propagation of excitation energy along the DNA molecule were studied, a new quasi particle, a proton exaction, was predicted, and its properties were studied.92 It turned out that the main contribution to the process of hot and cold spots of ultraviolet mutagenesis formation is made by the processes of propagation of excitation energy along the DNA molecule.65 All these results were used in the construction of a model for rare tautomeric forms of DNA bases formation upon irradiation of a DNA molecule with ultraviolet light.63 The rare tautomeric forms are stable when the respective bases are involved in cyclobutane thymine dimers.63 This is because the DNA strand bends once pyrimidine dimers arise, and the hydrogen bonds between the bases are broken between the bases that neighbor the cyclobutane pyrimidine dimers.93,94 The rare tautomeric forms of DNA bases are stable in DNA synthesis63 These conclusions were confirmed by experiments.77,78 The results of studies on the structure of the active centers polymerases show that the bases in rare tautomeric forms may exist in the active sites of polymerases77,78 These results served as the basis for the development of the polymerase-tautomeric models for targeted ultraviolet mutagenesis, radiation-induced bystander effects and radiation-induced genomic instability.60–80 Let us consider error-prone and SOS synthesis of DNA containing cis-syn cyclobutane cytosine dimmers CC and CC4*. We investigate to what biological consequences they can lead. In order to understand how delayed mutations can be formed, one must understand how DNA synthesis occurs, and how different DNA polymerases act.
The main function of DNA, as is known, is the preservation of hereditary information. Semi conservative replication provides preservation of hereditary information. Replication is the process of doubling DNA (DNA synthesis on a DNA template). Matrix synthesis, occurring during replication, repair, or transcription, follows the rules of complementarity of nitrogen bases (A-T and G-C), based on their particular chemical structure. As a result, DNA and RNA polymerases can accurately replicate the nucleotide sequence. Most of these enzymes strictly distinguish normal bases in matrix molecules; therefore, as a rule, chemical modifications of nucleotides in DNA lead to blocking of normal transcription and replication, that is, they are non coding lesions. As a rule, DNA synthesis is a highly accurate process. The accuracy of the synthesis is from 10–9 to 10–11 erroneous bases per pair of bases in the replication of the dividing cell.95 Damage to DNA is eliminated by a number of reparation mechanisms. These include photo reparation, excision repair, postreplicative repair, mismatch repair and others.96 If not all of the lesions is removed; it is possible to induce an error-prone or SOS system. In this case, DNA synthesis can also occur on DNA containing lesions. Such synthesis is called translesion synthesis; it can cause mutations.97 Mutations appear as a result of DNA polymerase errors. SOS replication and SOS repair of bacteria or error-prone replication and repair of mammals can lead to mutations as a result of the mechanism of the sliding clamp98 or by the operation of specialized DNA polymerases characterized by low synthesis accuracy.46,99 They act together and coordinated, so that the most suitable polymerase corresponding to this DNA damage is synthesized.46,99
Constitutive DNA polymerases
DNA polymerase is an enzyme involved in DNA replication. DNA polymerase synthesizes the daughter DNA strands during replication, they inserted canonical bases opposite damaged DNA sites during repair and therefore play a key role in genome reproduction and preservation of its primary structure. Human DNA polymerases δ and ε are key enzymes in the replication of chromosomes. Mutations in Pol δ in mice and humans lead to genomic instability, mutator phenotype and tumor genesis. The advent of genome sequencing techniques has identified damaging mutations in the proofreading domain of POLD1 as the underlying cause of some inherited cancers. Changes in expression or activity of POLD1 have been linked to senescence and aging.100 DNA polymerase III is the main polymerase that replicates DNA in E. coli. DNA polymerase III of E. coli and DNA polymerases δ and ε have 3¢®5¢-exonuclease activity performing the corrector function during DNA synthesis.101,102 It is known that DNA polymerase usually acts in combination with other enzymes and proteins. Enzymes and proteins involved in replication (there are more than 40 of them) are united into a single complex - replisome. The holoenzyme of the DNA polymerase can include the DNA polymerase itself, proteins regulating the rate of synthesis, 3'®5'-exonuclease, etc.101
Specialized DNA polymerases of mammals
To ensure efficient and timely replication of DNA molecules, organisms possess specialized DNA polymerases. Translesion synthesis is the process in which specialized DNA polymerases replicate opposite DNA lesions. There are several specialized DNA polymerases of eukaryotes, these are the zeta (Pol z), kappa (Pol κ), this (Pol η), theta (Pol θ), iota (Pol ι).103 By possessing a spacious preformed active site, these enzymes can physically accommodate a variety of DNA lesions and facilitate their bypass. Flexible DNA-binding domains and a variable binding pocket for the replicating base pair further allow these translesion polymerases to select specific lesions to bypass and favor distinct non-Watson-Crick base pairs.104
The Y family of DNA polymerases, including DNA polymerase eta (Pol η) play a key role in TLS. Mutations in the human POLH gene encoding Pol η underlie the genetic disease xero derma pigmentosa variant, characterized by sun sensitivity, elevated incidence of skin cancer, and at the cellular level, by delayed replication and hyper mutability after UV-irradiation. Pol η is a low fidelity enzyme when copying undamaged DNA, but can carry out error-free TLS at sites of UV-induced di thymine CPDs.105,106 The active site of Pol η has an open conformation that can accommodate CPDs, as well as cisplatin-induced intra strand DNA cross links. Pol η is recruited to sites of replication arrest in a tightly regulated process through interaction with PCNA.107 DNA polymerase kappa (Pol κ) is a specialized DNA polymerase that plays an important role in DNA damage tolerance through translation DNA synthesis.108 Pol ζ plays an important role in the protection of human cells by carrying out translation synthesis across bulky DNA adducts and cross-links.109
Specialized DNA polymerases of Escherichia coli
In order to deal with DNA damage and other stresses, Escherichia coli utilizes the SOS response, which regulates the expression of at least 57 genes. Y-family DNA polymerases are characterized by their specialized ability to copy damaged DNA in a process known as translation synthesis and by their low fidelity on “undamaged” DNA templates. Y-family polymerases exhibit various specificities for different types of DNA damage.110,111 In Escherichia coli, there are several specialized DNA polymerases, DNA polymerase IV and DNA polymerase V31 Pol IV and Pol V polymerases of the Escherichia coli are inducible components of the SOS system. All these DNA polymerases are part of a stress-induced process that allows them to function only when a high rate of mutation is beneficial to the body. DNA polymerases pols II, IV, and V perform the vast majority of translesion synthesis. Pol V can traverse a wide range of DNA lesions and performs the bulk of mutagenic translesion synthesis, whereas pol II and pol IV appear to be more specialized DNA polymerases.112 Depending upon the nature of the DNA damage and its sequence context, all three SOS-inducible DNA polymerases Pol II, Pol IV and Pol V, are involved in error-free and mutagenic translesion synthesis. For example, error-free and -1 frame shift translesion synthesis at a benzo(a)pyrene adduct requires both Pol IV and Pol V.113
Pol IV produces untargeted mutations, in addition, Pol IV is involved in the mutagenesis of non-dividing cells.114 Pol V carries out translesion synthesis to bypass a basic sites and thymine-thymine cyclobutane dimmers and TT (6–4)-photoproduct, especially TT (6–4)-photoproduct, in an error-prone manner.110,111 in vitro и in vivo.31,110 Very little replication past a T-T cis-syn cyclobutane dimmer normally takes place in Escherichia coli in the absence of DNA polymerase V.31,110 Pol V catalyses template-directed nucleotide incorporation and is responsible for most of the mutagenesis associated with the SOS response.115 Pol V contributes to untargeted mutagenesis.31 In non induced cells, Pol V is undetectable. In contrast, it is estimated that there are about 250 copies of Pol IV per cell. On SOS induction, it is believed that only about 15 molecules of Pol V are assembled per cell, whereas Pol IV levels reach approximately 2500 molecules.116 DNA polymerase IV appears much more frequently than Pol III. Approximately 250 Pol IV molecules account for 30 Pol III molecules.114 When DNA polymerase III is stopped, pol III is replaced by DNA polymerase IV capable of bypassing specific lesions.98 The actions of translesion polymerases are managed in part by ring-shaped sliding clamp proteins. Expression of translesion polymerases impedes cellular growth. Pol III-Pol IV switching represents a vitally important mechanism for regulating translesion synthesis in vivo by managing access of Pol IV to the DNA.117 The active form of DNA polymerase V is active mutasomal complex for translesion synthesis.118 Escherichia coli DNA polymerase V (pol V), a hetero trimeric complex composed of UmuD'2C, is marginally active. ATP and Rec A play essential roles in the activation of pol V for DNA synthesis including translesion synthesis.97,119,120 Several molecular mechanisms limit the amount of time that pol V has to access DNA.121
Sliding clamp mechanism
Most erroneously incorporated nucleotides are removed during DNA replication by 3¢®5¢ exonucleases. After making the error 3¢-ОН the end of the primer exits from the double helix DNA and the probability of dissociation of the DNA polymerase from the primer increases. DNA polymerase dissociates from DNA, after which it is associated with the exonuclease center of the same or another molecule.102,122 The rate of primer extension decreases by a factor of 103-106, depending on the particular combination of unpaired nucleotides. During this pause, erroneous nucleotides can be removed by the 3'®5'-exonucleases of this DNA polymerase, other DNA polymerases or autonomous 3'®5'-exonucleases. Removing the unpaired nucleotide, the 3'®5'-exonuclease is soon separated from DNA and polymerase synthesis resumes.102 The processiveness factor, i.e., subunit β, plays a decisive role in controlling the ratio between the polymerase and proofreading activities of DNA-polymerase III E. coli. Its molecules form a ring-like moving platform or a “sliding clamp” that contains a central hole for double-stranded DNA to pass through as the polymerase moves along the DNA. It positions DNA-polymerase III on the template and ensures the highly processive synthesis of DNA.102 A similar mechanism exists in mammals.
When a replicative DNA polymerase encounters a chemically altered base that stops synthesis, translesion synthesis takes place. The replicative polymerase is transiently replaced by a specialized or lesion bypass polymerase. Translesion synthesis involves the beta-clamp in prokaryotes and PCNA in eukaryotes. All DNA polymerases bind to the beta-clamp or PCNA. Pol II and Pol V can both extend replication intermediates formed when the replicative Pol III dissociates in the vicinity of the damage.122 A sliding β clamp works with all DNA polymerases E. coli.31,98 If the DNA is damaged, the damaged base on the leading strand stops the synthesis of DNA, which takes place with the help of Pol III. Synthesis of this DNA site continues with low-precision polymerases, such as Pol IV and Pol V, they bypass damage, after which Pol III can resume synthesis.31,98,123 Replacement of the polymerases occurs with the help of a sliding clamp mechanism. It simultaneously binds two different DNA polymerases, one of which is Pol III with a high accuracy of synthesis, and another specialized Pol IV polymerase or Pol II polymerase.123 Pol III-Pol IV switching represents a vitally important mechanism for regulating translesion DNA synthesis in vivo by managing access of Pol IV to the DNA.117 Proliferating cell nuclear antigen (PCNA) lies at the center of the faithful duplication of eukaryotic genomes and in stimulating DNA polymerases.124 DNA polymerase δ interacts with PCNA.125 Mammalian cells regulate the levels and activities of replicative and specialized polymerases to maintain genome integrity. Such regulation is quite intricate, and occurs at the transcriptional, post-transcriptional and translational levels. This is a more complex process than a single post-translational modification of PCNA. Numerous proteins converge on the Y-family polymerases to facilitate their recruitment.126 Conditions leading to slowed or stalled DNA replication cause replication stress. Replication stress contributes to genome instability.126 Replacing constitutive DNA polymerases with specialized DNA polymerases that are capable of synthesizing on a matrix containing lesions allow one to bypass damage and prevent cell death.106 Fine regulation, which makes it possible to involve the most suitable DNA polymerase at any given moment, ensures the highest possible synthesis accuracy in this situation. For example, specialized DNA polymerase pol ι is able not only to bypass UV photoproducts, but also to delay light-induced skin cancer.127 Thus, when an error-prone or SOS-system is induced, control for DNA bases is weakened, and even constitutive polymerase DNA Pol III E. coli and DNA polymerases δ and ε mammalian can incorporate bases opposite the cyclobutane dimmers. Even when an erroneous base pair is formed, the "sliding clamp" mechanism presses the DNA polymerase to the matrix, which prevents removal of the "wrong" base by the 3¢®5¢-exonucleases. In addition, DNA polymerases that generally do not have exonucleases (DNA polymerases IV or V E. coli or Pol ζ, Pol ι or Pol κ DNA polymerases) can involve in DNA synthesis. Moreover, specialized DNA polymerases can be pressed by a sliding clamp, which lowers the accuracy, but increases the synthesis rate, which, in the case of a large number of damages, helps to prevent cell death. Consequently, under error-prone and SOS-synthesis of DNA containing cyclobutane dimers, first, control over the bases of the template DNA is weakened. This, in particular, is expressed in the fact that DNA bases are inserted opposite cyclobutane dimers. Secondly, if an erroneous pair was formed, the "sliding clamp" mechanism presses the DNA polymerase to the template and does not allow the 3¢®5¢-exonucleases to remove the "improper base".
Polymerase-tautomeric model for targeted delayed base substitution mutations during error-prone or SOS synthesis of double-stranded DNA containing cis-syn cyclobutane cytosine dimmers CC4*
During SOS synthesis of DNA containing dimmers, nucleotide bases are inserted opposite the dimmers without the removal of the dimmer-containing sites. This is only possible when the DNA-polymerase is pressed on the DNA by the “sliding clamp”, obstructing the operation of exo nucleases, or when the synthesis involves specialized DNA polymerases, such as E. coli polymerase V or IV, or when the specialized DNA-polymerase is pressed on the DNA by the “sliding clamp”46,99 As the analysis of the operation of various DNA polymerases has shown, specialized and modified DNA polymerases incorporate canonical bases capable of forming hydrogen bonds with bases are part of cyclobutane pyrimidine dimers in template DNA.64 Seven new rare tautomeric conformations of G:C base pairs (Figure 2A–2H) are proposed that are capable of influencing the character of base pairing.52 Cis-syn cyclobutane cytosine dimers wherein a cytosine occurs in the rare tautomeric forms C1*, C2*, C5* or C6* were shown to cause targeted base substitution mutations only.42,52 Cis-syn cyclobutane thymine dimers wherein a thymine is in the rare tautomeric form C3* or C7* may result in targeted frameshift mutations (targeted insertions and targeted deletions).60 Let's see what biological consequences can lead cis-syn cyclobutane cytosine dimers CC4*.
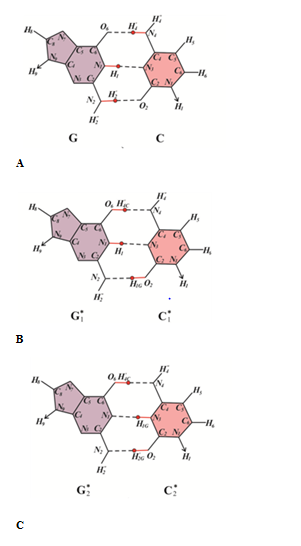

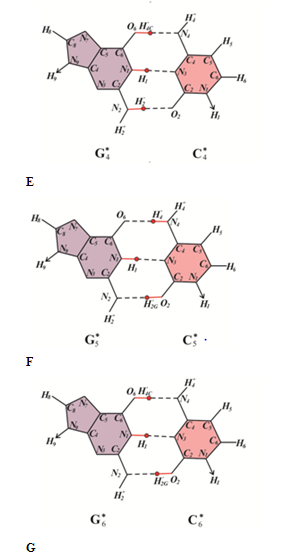
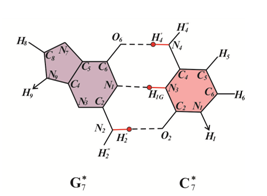
Figure 2 G: C base pairs formed by bases in canonical and rare tautomeric conformations. (A)(G) and (C) are in canonical tautomeric forms; (B-H) guanine (Gi*) and cytosine (Ci*) are in rare tautomeric forms, i=1¸7.
Let us examine the possible biological consequences of cis-syn CC4* cyclobutane cytosine dimers. In order to determine which of the canonical bases will be inserted by DNA-polymerases opposite cis-syn CC4* cyclobutane cytosine dimers, consider the constraints on the formation of hydrogen bonds between the C4* bases of the template DNA and the inserted bases (Figure 3) The rare C4* cytosine tautomer is capable of forming two hydrogen bonds with canonical guanine (Figure 3A). But C4* can form one hydrogen bond with canonical adenine (Figure 3B), one hydrogen bond with canonical cytosine (Figure 3C) and one H-bond with canonical thymine (Figure 3D). Consider a DNA site with a cis-syn CC4* cyclobutane cytosine dimmers (Figure 4A). Let other cis-sin cyclobutane pyrimidine dimers are located quite far from it. Since the damage is only one, the synthesis through the damage will go quite quickly and with high accuracy. For example, the synthesis will be carried out using Pol III DNA polymerase of Escherichia coli or eukaryotic DNA polymerase δ. If a wrong nucleotide is inserted opposite the cis-sin cyclobutane cytosine dimer CC4*, the erroneous nucleotides can be removed by 3'®5'-exonucleases. Therefore, with a high probability guanine will be inserted opposite cytosine C4* (Figure 4B). In this case the strand of DNA containing cis-sin cyclobutane cytosine dimer CC4* does not result in mutations (Figure 4C). So many cycles of DNA replication can continue. Mutations will not appear until the situation changes. Let a different, for example, canonical cyclobutane dimer form after a certain, perhaps long time, near the cis-syn of the cyclobutane cytosine dimer CC4* (Figure 5A). In this case, the synthesis will be less accurate. For example, the synthesis will continue to be carried out using DNA polymerase Pol III Escherichia coli or DNA polymerase δ eukaryotes, but in the presence of a sliding clamp. Then, if an erroneous base pair is formed, the sliding clamp will press the DNA polymerase to the DNA strand and prevent the 3'®5'-exonucleases from removing the “improper” base. Let us assume that in this case the control over the formation of pyrimidine-purine bases pairs only will remain. Consequently, canonical tautomeric forms of adenine can be incorporated opposite cytosine C4* with some probability (Figure 5B). In this case, C-G®T-A delayed targeted transition will result (Figure 5C).


Figure 3 Figure 1, Rare tautomeric state of cytosine C4* and structural analysis of pairing of cytosine C4* with canonical tautomer of DNA bases: (A) with guanine; (B) with adenine; (C) with cytosine; (D) with thymine.

Figure 4 Figure 2, Error-prone or SOS replication of the DNA containing CC and CC4* cis-syn cyclobutane cytosine dimers, when cis-syn cyclobutane cytosine dimers CC and CC4* do not lead to mutations: (A) a DNA site containing cis-sin cyclobutane cytosine dimers CC and CC4*; (B) guanine molecules are inserted opposite the rare C4* tautomeric form of cytosine and C-canonical tautomer of cytosine; (C) molecules of cytosine are inserted opposite molecules of guanine, mutations do not form.
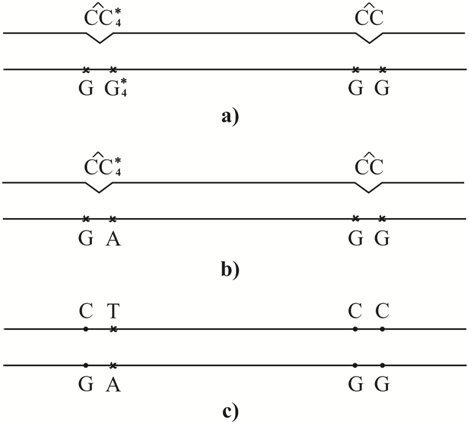
Figure 5 Figure 3, Error-prone or SOS-replication of the DNA containing CC and CC4* cis-syn cyclobutane cytosine dimers, when cis-syn cyclobutane cytosine dimmer CC4* result in C-G®T-A transition, but cis-syn cyclobutane cytosine dimmer CC do not result in mutation: (A) a DNA site containing cis-sin cyclobutane thymine dimers CC and CC4*; (B) a adenine is inserted opposite cytosine C4*, a molecules of guanine are inserted opposite molecules of canonical cytosine C; (C) a thymine is inserted opposite adenine, transition C-G®T-A is appeared.
Let, after some time after irradiation of the DNA with ultraviolet light, many other damages capable of stopping the synthesis of DNA appear near to the cis-syn cyclobutane cytosine dimer CC4*. Some of them can be caused, for example, by free radicals, the main cause of spontaneous mutagenesis. In (Figure 6), they are designated as Sp. Other DNA damages can be caused by the action of some other chemicals. As is well known, a large number of heavy metals and other chemicals have been detected in patients with cardiovascular and cancerous diseases.128 In (Figure 6A), they are designated as Ch. As experiments show, if there is a large amount of DNA damage, DNA polymerases with a lower rate and accuracy of synthesis are involved in the synthesis. For example, the synthesis will be carried out using DNA polymerases IV or V of Escherichia coli or some specialized DNA polymerase of eukaryotes. Perhaps they will be pressed by a sliding clamp. In this case, with a high probability, not only transitions, but also transversions can be formed. Canonical tautomeric forms of thymine can be incorporated opposite cytosine C4* (Figure 6B). In this case, G-C®T-A trans version will result. The insertion of a canonical tautomeric form of cytosine opposite cytosine C4* (Figure 6C) produces homologous C-G®G-C Trans version.

Figure 6 Figure 4, Error-prone or SOS-replication of the DNA containing CC and CC4* cis-syn cyclobutane cytosine dimers, when cis-syn cyclobutane cytosine dimmer CC4* result in C-G®A-T trans version or homologous C-G®G-C trans version, a canonical cis-syn cyclobutane cytosine dimmer CC result in C-G®A-T trans version: (A) a DNA site containing cis-syn cyclobutane cytosine dimmer CC and CC4*, as well as damages Ch. and Sp. capable of stopping the synthesis of DNA; (B) a thymine is inserted opposite cytosine C4* and canonical cytosine, which are part of the cis-syn cyclobutane cytosine dimmer CC4* and CC; (C) a cytosine is inserted opposite a cytosine C4*, a thymine is inserted opposite a canonical tautomer of cytosine C, which are part of the cis-syn cyclobutane cytosine dimer CC; (D) a adenine is inserted opposite a thymine, C-G®A-T trans version is formed; (E) a guanine is inserted opposite a cytosine, homologous trans version C-G®G-C is formed.
Polymerase-tautomeric model for targeted delayed base substitution mutations during error-prone or SOS synthesis of double-stranded DNA containing cis-syn cyclobutane cytosine dimers CC
Let's see if, under certain conditions, canonical cis-syn cyclobutane cytosine dimers result in any targeted mutations. This is a very important issue, as a rule, 88-95% of cis-syn cyclobutane pyrimidine dimers do not result in mutations.41 For this, we make a structural analysis and find out which canonical bases can form hydrogen bonds with canonical tautomeric form of cytosine. Of course, canonical tautomeric form of cytosine can form a pair with canonical tautomeric form of guanine (Figure 1A). The canonical tautomeric form of cytosine cannot form H-bonds with canonical tautomers of adenine Figure 1c or cytosine (Figure 1B) for steric reasons. But the canonical tautomeric form of cytosine can form hydrogen bonds with canonical tautomers of thymine (Figure 1C). This fact has long been known. Let's see under what conditions canonical cis-syn cyclobutane cytosine dimers can lead to mutations and find out what kind of mutations it can be. If there is only one or two cyclobutane cytosine dimers (Figure 4A), then translation synthesis will proceed fairly quickly and with high accuracy. DNA polymerase by pass a cis-syn cyclobutane cytosine dimer in a mostly error-free manner. Therefore, with a high probability guanine will be inserted opposite canonical cytosine C tautomer Figure 4b. In this case the strand of DNA containing cis-sin cyclobutane cytosine dimers CC does not result in mutations (Figure 4C). And so many cycles of DNA replication can continue.
Let a few other cyclobutane pyrimidine dimers form after a certain, perhaps long time, near the canonical cis-syn canonical cyclobutane cytosine dimer CC (Figure 5A). In this case specialized or modified DNA polymerase replicated past a cis-syn cyclobutane cytosine dimer with less accuracy. Let us assume that in this case the accuracy of control over the number of hydrogen bonds formed between the DNA bases will decrease. But control over the formation of pyrimidine-purine base pairs will continue. And in this case, guanine will be inserted opposite the canonical cytosine (Figure 5B) and mutations will not form (Figure 5C). Assume that after some time after DNA irradiation with ultraviolet light, near the canonical cis-sin cyclobutane cytosine dimer CC, many other damages capable of stopping the DNA synthesis appear. Some of them can be caused, for example, by free radicals, the main cause of spontaneous mutagenesis. In (Figure 6), I marked them as Sp. Other DNA damage can be caused by the action of some other chemical DNA.128 The main threats to human health from heavy metals are associated with exposure to lead, cadmium, mercury and arsenic. Long-term exposure to arsenic in drinking-water is mainly related to increased risks of skin cancer, but also some other cancers, as well as other skin lesions such as hyperkeratosis and pigmentation changes.129 In ref.130 metal concentrations were determined in tumor and healthy tissues of 101 head and neck cancer patients, using Atomic Absorption Spectrometry. The As, Cd, Cr, and Ni levels in tumor tissues were 3.4, 2.5, 1.3 and 1.5 times higher than those of healthy tissues (p<0.05), respectively. Mean blood levels of Cr and Ni in head and neck cancer cases (52.15 and 111.60μg/L, respectively) were significantly higher than those of controls (37.04 and 30.50μg/L, respectively).131 Passive smoking may interact with mutagen sensitivity and other risk factors to increase the risk of head and neck cancer.132 In (Figure 6), I marked them as Ch. As shown by experiments, if there is a large amount of DNA damage, DNA polymerases with lower speed and accuracy are involved in the translesion synthesis. In the case DNA polymerases replicated past a cytosine-cytosine cis-syn cyclobutane dimer is highly error-prone. Most likely, specialized DNA polymerase will be pressed by a sliding clamp. Only in this case transversions can form. Canonical tautomeric form of thymine may be formed opposite canonic tautomeric form of cytosine. Transversion С-G®А-T can form (Figure 6C).
The fact that under these conditions, mutations can appear opposite canonical cis-syn cyclobutane cytosine dimers, changes the situation dramatically. Let us estimate how many times the probability of occurrence of delayed base substitution mutations increases in this case in comparison with the case when only the dimers of СС4* are the source of such mutations. As a rule, only 5-12 % of cyclobutane dimers and (6-4) adducts result in replication errors.41 As shown in the polymerase-tautomer model of ultraviolet mutagenesis, these are cis-syn cyclobutane dimers with bases in rare tautomeric forms.60,61,64,68,69 Therefore, if all cis-syn cyclobutane dimers result in mutations, the probability of mutation formation (for this reason) will increase by 10-20 times. Seven new rare tautomeric conformations of the guanine and cytosine60 are proposed that are capable of influencing the character of base pairing. Only one of them can result in delayed base substitution mutations. Therefore, for this reason, the probability of delayed base substitution mutations will increase 70-140 times. It is interesting that the corresponding estimate for the canonical cis-syn cyclobutane thymine dimers gives an increase in the number of delayed targeted base substitution mutations 500-1000 times greater than if they were formed only opposite cis-syn cyclobutane thymine dimers of ТТ3*.75 Cis-syn cyclobutane thymine dimers are formed about 10 times more often than cis-syn cyclobutane cytosine dimers. Consequently, cis-syn cyclobutane thymine dimers will result in delayed targeted base substitution mutations approximately 70 times more often than cis-syn cyclobutane cytosine dimers. For targeted base substitution mutations appearing immediately after irradiation, the situation is different, namely, cis-syn cyclobutane cytosine dimers cause the targeted base substitution mutations 70 times more often than cis-syn cyclobutane thymine dimers. This is easily seen by studying the hot and cold spots of ultraviolet mutagenesis in.133 The formation of delayed targeted base substitution mutations caused by cis-syn cyclobutane thymine dimers was studied in Ref.75 It was concluded that with a very large number of DNA lesions, the targeted delayed base substitution mutations caused by cis-syn cyclobutane cytosine and thymine dimers will occur with similar probabilities. As we see, in fact, cis-syn cyclobutane thymine dimers will significantly more often cause delayed targeted base substitution mutations than cis-syn cyclobutane cytosine dimers.
Thus, cis-syn cyclobutane cytosine dimers СС4*, whose one or both bases are in such rare tautomeric forms that can form hydrogen bonds with both guanine and other canonical DNA bases, can be the source of the targeted delayed base substitution mutations. In addition, canonical cis-syn cyclobutane cytosine dimers CC can also result in targeted delayed base substitution mutations. Whether a delayed mutation will appear or not depends entirely on the presence or absence of other DNA damages near this damage. If there are no other DNA lesions or there are very few of them, then translesion synthesis will go quite accurately and mutations are not formed. If other lesions, capable of stopping the synthesis of DNA, are located next to the cis-syn cyclobutane cytosine dimer СС4* then the synthesis will involve other specialized DNA polymerases with a lower accuracy of synthesis. As a result, C-G®T-A transitions may appear. If there are a lot of damages, capable of stopping the synthesis of DNA, are located next to the cis-syn cyclobutane cytosine dimer СС4* or CC then the synthesis will involve specialized DNA polymerases with very low synthesis accuracy, in addition, their accuracy can be further reduced by the operation of the sliding clamp. In this case, the cis-syn cyclobutane cytosine dimer СС4* can result in C-G®A-T targeted delayed transversion of or C-G®G-C targeted delayed homologous transversion. But the canonical cis-syn cyclobutane cytosine dimer CC can result in the targeted delayed transversion of C-G®A-T only. New lesions may appear sometime after dimer formation. Only then will the conditions described above arise. And the cis-syn cyclobutane cytosine dimmer СС4* or CC can result in mutations through many cycles of replication after DNA has been damaged. Consequently, these are the delayed mutations.
The modern theories for mutagenesis cannot exhaustively explain many phenomena, including radiation-induced genomic instability and mechanism of delayed mutations formation. Usually, a polymerase paradigm of mutagenesis is used to explain them. The common accepted polymerase paradigm assumes that all cis-syn cyclobutane thymine dimers are identical, and the DNA polymerase is sometimes mistaken, sporadically inserting uncomplimentary bases opposite to cyclobutane pyrimidine dimers. However, such approach contradicts a lot of the experimental facts. I proposed and develop polymerase-tautomeric models for ultraviolet mutagenesis, radiation-induced bystander effects and radiation-induced genomic instability. Polymerase-tautomeric model for mechanism of targeted delayed base substitution mutations caused by cis-syn cyclobutane cytosine dimers is proposed. Targeted delayed mutations are mutations that can appear after several cycles of replication after the mutagen has been exposed to damage that can stop the synthesis of DNA. They can be caused, in particular, by ultraviolet light. Ultraviolet irradiation can lead to a change in the tautomeric states of DNA bases. Cytosine can form seven rare tautomeric forms, which are stable if the related nucleotides are components of cyclobutane dimers. Structural analysis of the insertion of the bases showed that opposite rare tautomeric form of cytosine С4* guanine can be incorporated, but may be inserted any other canonical base so that between them hydrogen bonds are formed. If in the synthesis of DNA containing the cis-syn cyclobutane cytosine dimer CС4*, involved DNA polymerases with relatively high fidelity of synthesis, mutations not appear. However, if further DNA synthesis will involve DNA polymerases having a low fidelity of synthesis, there may be base substitution mutations. Moreover, they may be formed through many cycles of replication after DNA has been damaged. Consequently, these are the delayed mutations. The cis-syn cyclobutane cytosine dimer СС4* can result in targeted delayed transitions C-G®T-A, targeted delayed transversion C-G®A-T of or targeted delayed homologous transversion C-G®G-C. In addition, it was found that even canonical cis-syn cyclobutane cytosine dimers can result in targeted delayed base substitution mutations. Opposite canonical thymine cytosine can be incorporated only. Canonical cis-syn cyclobutane cytosine dimers CC may result in targeted delayed trans versions C-G®A-T only. Such mutations can be formed only in the case where next to the canonical cis-syn cyclobutane cytosine dimer there is a lot of other DNA damage. It was concluded that the reason for the instability of the genome is a large amount of DNA damage. Not all these damages must necessarily be mutagenic. If these lesions are capable of stopping DNA synthesis, then they can lead to translesion synthesis, involved DNA polymerases with a low synthesis accuracy and, therefore, contribute to mutagenesis. As was shown earlier, cis-syn cyclobutane cytosine dimers can be of seven types, depending on which rare tautomeric forms are the DNA bases containing in the cis-syn cyclobutane cytosine dimers. Each of these types of dimers can lead to certain types of mutations. Seven new rare tautomeric conformations of G:C base pairs (Figure 2) are proposed that are capable of influencing the character of base pairing.52 Cis-syn cyclobutane cytosine dimers wherein a cytosine occurs in the rare tautomeric forms C1*, C2*, C5* or C6* were shown to cause targeted base substitution mutations only.42,52 Cis-syn cyclobutane cytosine dimers wherein a cytosine is in the rare tautomeric form C3* or C7* may result in targeted frame shift mutations (targeted insertions and targeted deletions).60 The cis-syn cyclobutane cytosine dimer СС4* or CC can result in targeted delayed base substitution mutations.
If the canonical cis-syn cyclobutane cytosine dimers lead to the mutations, the probability of formation of the targeted delayed base substitution mutations will increase by 70-140 times. Cis-syn cyclobutane thymine dimers will result in delayed targeted base substitution mutations approximately 70 times more often than cis-syn cyclobutane cytosine dimers. This is a very interesting conclusion, considering that cis-syn cyclobutane cytosine dimers result 50-100 times more often in targeted base substitution mutations that appear immediately after irradiation than cis-syn cyclobutane thymine dimers. Radiation-induced genome instability and radiation-induced bystander effects, in particular under the influence of long-wave ultraviolet light (UVA), are often explained by oxidative damage to DNA bases.48 However, for example, in Ref.15 oxidative damage was not found at all. At the same time, pyrimidine cyclobutane dimers and 6-4 adducts range from 78% to 84% of all lesions in long-wave irradiation of DNA.24,25 In addition, any mutations that appear opposite cyclobutane pyrimidine dimers or 6-4 adducts can be explained easily and naturally using a polymerase-tautomeric model of mutagenesis.61,64,67–69 Thus, the polymerase-tautomeric models are able to explain targeted base substitution mutations,61,64 targeted insertions,61,68 targeted deletions,61,69,70 targeted complex insertions61,70 and targeted delayed base substitution mutations,72,75 and the causes of the formation of hot and cold spots of ultraviolet mutagenesis.65 In addition, it is able to explain such radiation-induced bystander effects as untargeted base substitution mutations and untargeted insertions.62, 69,73,74 It is able to explain such phenomena of genomic instability as the targeted delayed base substitution mutations72,75 To explain the radiation-induced bystander effects and radiation-induced genomic instability, there is no need to change the paradigm of radiation biology or genetics. It is enough just to change the paradigm in mutagenesis. Several works have been devoted to testing the ideas of Watson & Crick77–79 It is observed a dGTP/dT77 and a dCTP/dA mispairs78 with one of the bases in rare tautomeric forms in the active site of DNA polymerases. These structures unambiguously77,78 demonstrate that tautomeric base pairs can form in the polymerase active site, providing strong support of the rare tautomer hypothesis and polymerase-tautomeric models of the author through direct structural evidence.
None.
None.

©2018 Grebneva. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.