International Journal of
eISSN: 2573-2889


Research Article Volume 1 Issue 1
Donetsk Physical and Technical Institute, Ukraine
Correspondence: Helen A Grebneva, Donetsk Physical and Technical Institute, NAS of Ukraine, 03680 Kiev, Ukraine
Received: September 12, 2016 | Published: November 10, 2016
Citation: Grebneva HA. A polymerase tautomeric model for targeted substitution mutations formation during error prone and SOS replication of doublestranded DNA containing Cis-syn cyclobutane cytosine dimers. Int J Mol Biol Open Access. 2016;1(1):4-17. DOI: 10.15406/ijmboa.2016.01.00002
A polymerase-tautomer model for ultraviolet mutagenesis is described that is based on formation of rare tautomeric bases in cis-syn cyclobutane pyrimidine dimers. The model is based on that fact that during error-prone and SOS synthesis the induced DNA polymerase inserts canonical bases opposite the dimers in a complementary way in contrast to un complementary one in the conventional models. There are same types of potential mutagens damages in cis-syn cyclobutane cytosine dimers. They correspond to 7 basic types of rare tautomeric conformations of cytosine. Error-prone and SOS replication of double-stranded DNA having cis-syn cyclobutane cytosine dimers, with one or both bases in 4 basic types of rare tautomeric conformations of cytosine (the dimers CC1*, CC,2*, CC5* and CC6*, with the *indicating a rare tautomeric base and subscript referring to the particular conformation), results only in targeted substitution mutations. The structural analysis indicates that three types of cis-syn cyclobutane cytosine dimer containing a single tautomeric base (the dimers CC1*, CC2*, CC2*¢) can cause G: C→A: T transition or G: C→C: G homologous transfusion. Another two dimers (CC5* and CC6*¢¢) can result only in G: C→Т: A transverse. The dimer CC6* can cause G: C→A:T transition, G:C→C:G homologous transverse or G:C→T:A transfusion. If both bases in the dimer are in a rare tautomeric form, then tandem mutations can be formed. The 90 substitution mutations have been interpreted from the point of view of polymerase-tautomer model of UV mutagenesis.
Keywords: UV-mutagenesis, Rare tautomeric forms, Targeted transitions, Targeted transversions, Cis-syn Cytosine cyclobutane dimers, Error-prone replication, SOS replication
Features of UV mutagenesis
Nearly the 48 agents were already known to induce somatic-cell mutations.1 They result in cancer, aging, diabetes, and other chronic diseases.2 Exposure of male mice to genotoxic agents can increase mutation frequencies in their unexposed descendants. This phenomenon, known as transgenerational genomic instability, can persist for several generations. However, little is known about the underlying mechanism.3 Delayed mutations and untargeted mutations are two features of genomic instability.4,5 In recent decades untargeted and delays mutations are combined in bystander effects.6-9 The bystander effect is defined as the induction of cellular damage in un irradiated cells, induced by irradiated cells in the surrounding area.7 There are now known to be many late expressed effects of exposure that cannot simply be explained on the basis of direct ionizing radiation DNA damages. Examples include genomic instability, bystander effects and adaptive responses.6-9
As to understand the mechanisms of the untargeted and delayed mutations formation you first need tounderstand themechanisms ofthe targeted base substitutions, deletions, insertions, and complex frame shift mutations. In order to understand the mechanisms of mutations formation by various mutagens, you first need to develop a model of these mechanisms caused by any one specific mutagen. Ultraviolet mutagen is understood better than anyone. So it is need to understand of mechanisms formation of different mutations that appear when DNA molecule is irradiated with UV light.
Ultraviolet (UV) radiation produces cyclobutane pyrimidine dimers and pyrimidine-pyrimidone (6-4) photoproduct from (78% to 84%),10,11 hydrated bases (1%-7%) (cytosine bases form more often), linked DNA-protein and DNA-DNA, and break in filaments (less than 1%) in Escherichia coli ,12,13 yeast,14 in xeroderma pigmentosum variant cell extracts and in mammalian cells under induced by UVB irradiation.15 In addition to pyrimidine-pyrimidine adducts, the purines-pyrimidine adducts are formed, for example, in consecution TpA, but they form very rarely.16 Cis-syn cyclobutane pyrimidine dimers account for a large majority of UV-induced mutations.17,17 If not all dimers are moved cyclobutane pyrimidine dimers19-22 and pyrimidine-pyrimidone (6-4) photoproduct 23,24 may produce mutations. Replication past the cis-syn T-C cyclobutane dimer, like replication past its T-T and U-U counterparts, is in fact >95% accurate and that the frequencies of bypass are also very similar for these photoproducts.25 Overall, 5 - 12 % of cyclobutane dimers and (6-4) adduct result in replication errors.26 Cis-syn cyclobutane pyrimidine dimers constitute a much more important premutagenic lesion than (6-4) pyrimidine-pyrimidone photoproducts, and it has been estimated that cis-syn cyclobutane pyrimidine dimers account for ≈80% of UV-induced mutations in mammalian cells.15 In addition to the formation of cyclobutane dimers at TT sites, UV induces the formation of cyclobutane dimers at 5′-TC-3′ and 5′-CC-3′ dipyrimidine sites, and both in yeast and humans, UV-induced mutations occur predominantly by a C-to-T transition at the 3′ base.27-29 Usually mutations occur opposite the photoproducts during error-prone and SOS replication, repair or transcription.19,25,30-36 Such mutagenesis is termed targeted.37,38 Sometimes mutations are formed in the vicinity of the damage, a process that is termed untargeted mutagenesis.39,40 Sometimes delayed mutations are formed 41-44. Untargeted and delayed mutations are two features of genomic instability.5 Cyclobutane pyrimidine dimers and (6-4) photoproducts cause substitution mutations (transitions and transversions),25,45,46 frame shift mutations (deletions and insertions)39,47-49 and complex frame shift mutations.50,51 Mismatch-repair systems suppress only about half of UV mutations.52 Delayed mutation and untargeted mutations are two features of genomic instability.5 Sunlight ultraviolet irradiation has been implicated in the etiology of human skin cancer.53 As a rule sewed DNA-protein and DNA-DNA and break in filaments result only in frame shift mutations. Cytosine dimers more often result in mutations then thymine dimers. Hot spots of UV-mutagenesis are sites consisting of photoproducts having thymine and cytosine bases or two cytosine bases.27,28
DNA polymerases involved in SOS synthesis of DNA
As a rule, DNA synthesis is a highly accurate process. If an erroneous base is inserted during the replication process, it is usually removed by 3¢→ 5¢exonuclease activity.54,55 UV irradiation generates cyclobutane dimers and 6-4 adducts in DNA, which if not removed by excision repair, result in the induction of the error-prone or SOS system. Error-prone or SOS induction allows DNA synthesis to occur even on templates containing cyclobutane dimers and 6-4 adducts;56 replication on a damaged DNA template, however, results in mutations. Specialized mutagenic DNA polymerases E. coli poll II, poll IV, and poll V are capable of replicating past DNA lesions, a process called translation synthesis.57-59 Lesion bypass during in vitro replication of duplex DNA containing cis-syn cyclobutane thymine dimers occurs by translation synthesis.60 Translation replication is carried out by specialized DNA polymerases.61 Mutations arise when modified by sliding clamp62,63 or specialized low fidelity62,64-74 DNA polymerases are involved in DNA synthesis. There is a cooperative and sequential assembly of translation synthesis polymerases in response to DNA damage.75
Poll IV and Poll V polymerases of the Escherichia coli are inducible components of the SOS system.39,76-79 They have low-fidelity DNA polymerase activity.80 Poll IV and Poll V appear to extend mismatched base pairs efficiently in the absence of other proteins. Polymerase Poll V catalyses template-directed nucleotide incorporation. Poll IV produces untargeted mutations.39,78 Poll V, in contrast to Poll III, does not possess any associated proof-reading functions.76,77 This mutator activity is influenced by the defective proof-reading sub-unit of Poll III.79,81
DNA polymerase poll V is responsible for most of the mutagenesis associated with the SOS response.82,83 Escherichia coli DNA polymerase V bypasses cis-syn-cyclobutane thymine-thymine dimer and TT (6-4)-photoproduct, especially TT (6-4)-photoproduct, in an error-prone manner.74 DNA polymerase poll V bypasses T-T cis-syn cyclobutane dimers in vitro and in vivo.74,84-86 Very little replication past a T-T cis-syn cyclobutane dimer normally takes place in Escherichia coli in the absence of DNA polymerase V.
DNA polymerases involved in error-prone synthesis of DNA
Poll η and Poll ζpolymerases of the yeast are inducible components of the error-prone system.65-68 In the case of yeast Poll ζ is highly error-prone.65,66,70,71,87 DNA polymerase ζ replicated past a thymine-thymine cis-syn cyclobutane dimer.68 DNA polymerase ζ is responsible for insertion in bypass of and a basic site, T-T (6-4) photo adduct and T-T cis-syn cyclobutane dimer events other than those in which polymerase η performs this function.68 The Poll ζ translation synthesis DNA polymerase is responsible for over 50% of spontaneous mutagenesis and virtually all damage-induced mutagenesis in yeast.88 Poll ζ catalyzes nucleotide incorporation opposite AAF-guanine and TT (6-4) photoproduct with a limited efficiency. Secondly, more efficient bypass of these lesions may require nucleotide incorporation by other DNA polymerases followed by extension DNA synthesis by Poll ζ.89
DNA polymerase η was found to be involved only rarely in the bypass of the T-T (6-4) photo adduct or the a basic sites in the sequence context used, although, as expected, it was solely responsible for the bypass of the T-T cis-syn cyclobutane dimer.68 The human DNA polymerase pollη bypasses cis-syn-cyclobutane thymine-thymine dimer efficiently in a mostly error-free manner but does not bypass TT (6-4)-photoproduct.90 Drosophila DNA polymerase pollη efficiently bypass a cis-syn cyclobutane thymine-thymine dimer in a mostly error-free manner. Drosophila DNA polymerases pollη shows limited ability to bypass a (6-4)-photoproduct at thymine-thymine (6-4)-photoproduct or at thymine-cytosine (6-4)-photoproduct in an error-prone manner.90 Pollη plays the protective role of against UV-induced lesions and the activation by UV of pollη-independent mutagenic processes.91 Although accessory proteins clearly participate in pollη functions in vivo, they do not appear to help suppress UV mutagenesis by improving pollη bypass fidelity per se.92 The yeast DNA polymerase η bypasses a cis-syn cyclobutane pyrimidine dimers and (6-4) photoproducts efficiently and accurately.93 DNA polymerase η plays an important in vivo role in inserting G opposite the 3′ T of 6-4 TT photoproducts.94
DNA polymerase has been implicated in translation DNA synthesis of oxidative and UV-induced lesions. Drosophila DNA polymerase pollι efficiently bypass a cis-syn cyclobutane thymine-thymine dimer in a mostly error-free manner.89 Purified human Pollι responded to a template TT (6-4) photoproduct by inserting predominantly an A opposite the 3′ T of the lesion before aborting DNA synthesis. In contrast, human Poll was largely unresponsive to a template TT cis-syn cyclobutane dimer.95 In cells lacking Poll is responsible for the high frequency and abnormal spectrum of UV-induced mutations, and ultimately their malignant transformation.73 The mutagenic properties of a lesion can depend strongly on the particular enzyme employed in bypass.84 Human DNA polymerase κ (Pollκ) is unable to insert nucleotides opposite the 3′T of a cis-syn T-T dimer, but it can efficiently extend from a nucleotide inserted opposite the 3′T of the dimer by another DNA polymerase.96 Human mitochondrial DNA polymerase pollγ mis incorporated a guanine residue opposite the 3'-thymine of the dimer only 4-fold less efficiently than it incorporated an adenine.72 De Marini97 shown that different mutagens induce the same primary class of base substitution and frameshift mutations in most organisms, this reflecting the conserved nature of DNA replication and repair processes.
Sliding clamp in mutagenesis
DNA polymerase III (Poll III) plays the main role in SOS replication in bacterial cells.81,98 UV-induced mutagenesis has been studied extensively in E. coli, and several E. coli DNA-polymerases involved in the process have been identified. Most erroneously incorporated nucleotides are removed during DNA replication by 3¢→ 5¢exonuclease, which are components of DNA-polymerases I and II, or by autonomous 3¢→ 5¢exonuclease (subunit ε of DNA-polymerase III).81 The possessiveness factor, i.e., subunit β, plays a decisive role in controlling the ratio between the polymerase and proofreading activities of DNA-polymerase III.99 Its molecules form a ring-like moving platform or a “sliding clamp” that contains a central hole for double-stranded DNA to pass through as the polymerase moves along the DNA. It positions DNA-polymerase III on the template and ensures the highly processive synthesis of DNA.100 Even when an erroneous base pair is formed, the sliding clamp mechanism presses the DNA polymerase against the template and prevents the 3¢→5¢-exonuclease from removing the “improper” base, this result in mutations.
DNA polymerases δ (Pollδ) and ε (Pollε) play the main role in error-prone replication in eukaryotic cells [101,102]. Mammalian proliferating cell nuclear antigen (PCNA) is the processivity factor in sliding clamp for essential eukaryotic DNA polymerases δand ε. The trimetric PCNA ring is striking similar to the dimeric ring formed by the β subunit (processivity factor) of the essential Escherichia coli DNA polymerase III holoenzyme and the gene 45 protein of T4 phage.99,103 Even when an erroneous base pair is formed, the sliding clamp mechanism presses the DNA polymerase against the template and prevents the 3¢→5¢-exonuclease from removing the “improper” base, this result in mutations. Cyclobutane pyrimidine dimers are responsible for most of the PCNA ubiquitin tion events after UV-irradiation.104
Sliding clamps are used by numerous different DNA polymerases and repair proteins in both prokaryotic (β clamp)105 and eukaryotic (PCNA clamp) organisms.106 Sliding β clamps functions with Poll I, Poll III holoenzyme and with the three damage inducible polymerases Poll II, Poll IV, and Poll V.62,74 During DNA damage, a damaged base on the leading strand will halt fork progression by the Poll III. A low-fidelity polymerase like Poll IV and Poll V presumably trade places with the stalled Poll III on β for lesion bypass, after which the Poll III may resume synthesis.107,108 DNA polymerases Poll III, Poll IV and Poll V increase cell fitness.109 The β sliding clamp of E. coli binds two different DNA polymerases at the same time. One is the high-fidelity Poll III chromosomal replicas and the other is Poll IV or Poll II polymerase.107,110
When the error-prone or SOS system is induced, control over the templating of bases becomes weaker, and bases are inserted opposite dimers. Even when an erroneous base pair is formed, the “sliding clamp” mechanism presses the DNA-polymerase against the template and prevents the 3¢→ 5¢exonuclease from removing the “improper” base. Alternatively, synthesis is performed by DNA polymerases having no exonuclease at all (e.g., E. coli DNA polymerase IV or V, or DNA polymerase Pollζ, Pollι or Pollκ).
DNA polymerase incorporates canonical bases capable of forming hydrogen bonds with cyclobutane pyrimidine dimers in template DNA
The efficiency and fidelity of nucleotide incorporation by high-fidelity replicative DNA polymerases are governed by the geometric constraints imposed upon the nascent base pair by the active site. Kinetic analyses of nucleotide incorporation opposite a cis-syn cyclobutane thymine-thymine dimer and an identical nondamaged sequence by yeast DNA polymerase η (Pollη) strongly support a mechanism in which the nucleotide is directly inserted opposite the cis-syn cyclobutane thymine-thymine dimer by using its intrinsic base-pairing ability without any hindrance from the distorted geometry of the lesion.66 Both a human polymerase κ (Pollκ) and polymerase η rely on Watson-Crick hydrogen bonding. A low-fidelity polymerase η shows more of a dependence upon Watson-Crick hydrogen bonding than doe’s human Pollκ, which differs from Pollηin having a higher fidelity.111 These and other results support conclusion that during the error prone or SOS synthesis, the canonical bases, which can form hydrogen bonds with bases of the template DNA, are incorporated opposite cyclobutane pyrimidine dimers.112
Models of mutagenesis
Polymerase paradigm of mutagenesis: At present, the conventional paradigm relates the reason of mutations exclusively to sporadic errors of DNA polymerases.74,82,83,113,114 Bresler113 proposed a mechanism for the formation of base-substitution mutations during the synthesis of DNA containing cyclobutane pyrimidine dimers. He assumed that the mutations arise because the DNA-polymerase sometimes incorporates non complementary nucleotides opposite the cyclobutane pyrimidine dimers. From the point of view of mutagenesis, this model assumes that, for example, all the cyclobutane thymine dimers are identical, and that mutations are induced by errors produced by DNA-polymerases when using DNA containing dimers as a template. However the approach resting exclusively on the polymerase paradigm is limited, it contradicts several experimental facts and can’t explain some mutagenesis phenomena.112
The reasons of mutation origination are explained by the so-called “A-rule”.113,116 The experiment shows that adenine (A) is most frequently incorporated opposite T (cis-syn) T dimers (94 %), guanine (G) - opposite T (cis-syn) C dimers (95%).25,26,66,68, 74,93,116-118 Poll V incorporates adenine opposite T (cis-syn) T dimers (98 %).74 Pollη incorporates adenine opposite T (cis-syn) C dimers (99 %) and guanine (G) opposite T (cis-syn) C dimers.46,66,73,117,118 Pollηи Polleta incorporates guanine opposite T (cis-syn) C [93]. Polymerase exo-T7 DNA poll incorporates adenine opposite T (cis-syn) T dimers (98 %).116 Polymerases pollη and poll V incorporates adenine opposite 3¢-T TT.93,119 In vitro replication studies of Pollιshow that it replicates past 5'T-T3' and 5'T-U3' cyclobutane pyrimidine dimers, incorporating G or T nucleotides opposite the 3' nucleotide.73 Pollη incorporates G opposite the 3'T or (6-4) adducts.94 Cyclobutane pyrimidine dimers as well as, to a lesser extent, the thymine-thymine pyrimidine-pyrimidone (6-4) photo product, were bypassed. Poll βmostly incorporates the correct dATP opposite the 3'-terminus of both cyclobutane pyrimidine dimers CPD and the (6-4) photoproduct but canals misinsert dCTP.120 Purified yeast Pollζperformed limited translation synthesis opposite a template TT (6-4) photoproduct, incorporating A or T with similar efficiencies (and less frequently G) opposite the 3' T, and predominantly A opposite the 5' T.89 DNA Pollζ,essential for UV induced mutagenesis, efficiently extends from the G residue inserted opposite the 3' T of the (6-4) TT lesion by Pollη, and Pollζ inserts the correct nucleotide A opposite the5' T of the lesion.118 These biochemical observations are in concert with genetic studies in yeast indicating that mutations occur predominantly at the 3' T of the (6-4) TT photoproduct and that these mutations frequently exhibit a 3' T→C change that would result from the insertion of a G opposite the 3' T of the (6-4) TT lesion. It has been shown that a cytosine or an adenine is the predominant nucleotide inserted opposite abasic sites.121 Basing on polymerase paradigm Taylor conclude that “the instructional or non-instructional behavior of a lesion in directing nucleotide insertion is not an invariant property of the lesion, but depends on the structure and mechanism of the polymerase involved”.82
These models undoubtedly describe some cases and features of mutagenesis. In E. coli, DNA polymerases IVandV, as a rule, function when DNA polymerase III has a defective ε subunit.63,81,108 However, the majority of mutations occur when DNA polymerase III has fully functional 3¢→ 5¢ exonuclease proofreading activity (a fully functional ε subunit), when the SOS system is induced, and when the “sliding clamp” mechanism is operating.56,59 Hence, this generally accepted paradigm describes only a small part of mutagenesis in E. coli. Moreover, this approach ignores the structure of the DNA damage.
Tautomeric model of mutagenesis: In the paper by Watson and Crick122 it was stated
that the spontaneous mutagenesis is based on capability of nucleotide bases to change their tautomeric state, which influences the character of base pairing. The participation of rare tautomeric forms in mutagenesis was repeatedly discussed.123-125 The role of tautomeric transitions in mutagenesis of the analogues of the bases is almost universally recognized.126 The mutagenesis models resting on rare tautomer hypothesis are, in fact, physical-chemical. Usually, they are simply stating that a change in the tautomeric state of bases is possible and concluding an erroneous pairing and, consequently, base substitution mutation to result from that change. The biological data are, as a rule, ignored. Spontaneous mutagenesis models suppose that the tautomeric state of DNA bases can be changed due to thermal vibrations. However, such bases in rare tautomeric forms will, most likely, be effectively removed by repair systems. The operation of free radicals formed in metabolism processes is known to be the main cause of spontaneous mutagenesis. These free radicals give damages of bases, sugar-phosphate stoma, etc.127,128 The removal of DNA structure damages by repair systems and conditions for them to be a template for DNA synthesis, etc, are not usually examined. Wang et al. provides structural evidence for the rare tautomer hypothesis of spontaneous mutagenesis.129
There are now two practically independent approaches to explaining mutagenesis. Physicists and chemists consider polymerases to be important; however, they largely ignore their role in the process and rely almost exclusively upon Watson and Crick’s paradigm.123,130,131 In contrast, most biologists cite Watson and Crick’s idea, but base their research on the polymerase paradigm.74,82,83,113,114
Bystander Effects: Bystander effects include untargeted and delays mutations.6-9 Naga
Saw and little first reported bystander effects.132 It is believed now that untargeted and part of delayed mutations appears on not damaged DNA sites.5 These, so called, untargeted effects are demonstrated in cells that are the descendants of irradiated cells either directly or via media transfer (radiation-induced genomic instability) or in cells that have communicated with irradiated cells (radiation-induced bystander effects).133 The dogma that genetic alterations are restricted to directly irradiated cells has been challenged by observations in which effects of ionizing radiation, characteristically associated with the consequences of energy deposition in the cell nucleus, arise in non-irradiated cells. These, so called, untargeted effects are demonstrated in cells that have received damaging signals produced by irradiated cells (radiation-induced bystander effects) or that are the descendants of irradiated cells (radiation-induced genomic instability).134
The most convincing explanation of radiation-induced genomic instability attributes it to an irreversible regulatory change in the dynamic interaction network of the cellular gene products, as a response to non-specific molecular damage.135 The central role of cell-cell communication on non-targeted effects is underlined.136 The discovery of non-targeted and delayed radiation effects has challenged the classical paradigm of radiobiology.137 It is assumed that a radiation cancer-causing target is protein.138 Furthermore, it is still not known what the initial target and early interactions in cells are that give rise to non-targeted responses in neighboring or descendant cells.6 Moreover,itwasconcludedthat only a third of the variation in cancer risk among tissues is attributable to environmental factors or inherited predispositions. The majority is due to “bad luck,” that is, random mutations arising during DNA replication in normal, noncancerous stem cells.139
Deamination of cytosine in mutagenesis: Cytosine easily undergoes hydrolytic Deamination.140 One hypothesisis that UV-induced mutations occur only after deamination of the cytosine or 5-methylcytosine within the pyrimidine dimer.141 Two models have been proposed: "error-free" bypass of deaminated cytosine-containing cyclobutane pyrimidine dimers by DNA polymerase η, and error-prone bypass of cyclobutane pyrimidine dimers and other UV-induced photo lesions by combinations of translation DNA synthesis and replicative DNA polymerases the latter model has also been known as the two-step model, in which the cooperation of two (or more) DNA polymerases as misinserters and (mis)extenders is assumed.142 Sunlight-induced cytosine to thymine mutation hotspots in skin cancers occur primarily at methylated CpG sites that coincide with sites of UV-induced cyclobutane pyrimidine dimer formation. The cytosine or 5-methyl-cytosine in cyclobutane pyrimidine dimers are not stable and deaminate to uracil and thymine, respectively, which leads to the insertion of adenine by DNA polymerase η and defines a probable mechanism for the origin of UV-induced cytosine to thymine mutations.143,144 There is evidence to suggest that, depending on solvent polarity, a cytosine or a 5-methylcytosine in a cis-syn cyclobutane pyrimidine dimer can adopt three tautomeric forms, one of which could code as thymine.145 Some data implicate the deamination of cytosine to uracil as a possible cause, but other results appear to indicate that the rate of deamination is too low for this to be significant in Escherichia coli .25
Polymerase-tautomeric model for ultraviolet mutagenesis: I have attempted to Construct a polymerase-tautomeric model for UV-induced mutagenesis,112,146-163 based on idea by Watson and Crick122 that changes in tautomeric state are possible for DNA bases. A mechanism for changes in the tautomeric state of base pairs has been proposed.146,152,161-163 It was assumed that the tautomeric state of the constituent bases may change during the formation of cyclobutane pyrimidine dimers.121,146,152 A mechanism for changes in the tautomeric state of base pairs has been proposed for the case when DNA is UV-irradiated and cyclobutane pyrimidine dimers are formed.149,151 Five new rare tautomeric conformations of the adenine and thymine150,152 and seven of the cytosine and guanine146 are proposed that are capable of influencing the character of base pairing. It is shown that such rare tautomeric forms of DNA bases are stable when they are parts of the cis-syn cyclobutane pyrimidine dimers and they are stable under DNA synthesis.112
Cis-syn cyclobutane thymine dimers wherein a thymine occurs in the rare tautomeric forms T1*, T4*, or T5* were shown to cause targeted base substitution mutations only.112,149 Cis-syn cyclobutane thymine dimers wherein a thymine is in the rare tautomeric form T2* may result in targeted frame shift mutations (targeted insertions and targeted deletions).156,157 I propose the mechanisms of targeted insertions formation during error-prone or SOS synthesis of DNA containing cis-syn cyclobutane cytosine155 and thymine156 dimers. A mechanisms was proposed for targeted complex insertions158 and targeted deletions157,159 caused by cis-syn cyclobutane thymine dimers. Cis-syn cyclobutane thymine dimers wherein a thymine is in the rare tautomeric form T3* may result in targeted delayed base substitution mutations.160 Polymerase-tautomeric model for untargeted substitution mutations formation when DNA molecule contains cis-syn cyclobutane thymine dimers also have been developed.148,151,154 A polymerase-tautomeric model was developed for the formation of hot and cold spots of UV-induced mutagenesis.153
Mechanism of alteration of DNA bases tautomeric state under cis-syn cytosine dimers formation
I use the polymerase-tautomer model for ultraviolet mutagenesis to determine which targeted mutations form by the error-prone or SOS replication opposite cis-syn cyclobutane cytosine dimers. The polymerase-tautomer model for ultraviolet mutagenesis is based on formation of rare tautomeric bases in cis-syn cyclobutane pyrimidine dimers. It was found that cytosine, guanine and other nucleic acid bases primarily exist in the form of two tautomer canonic and hydroxo forms with a possible small presence of the amino tautomeric form.124,125,164 The existence of up to four tautomers of guanine, namely enol-N7H, keto-N7H, keto-N9H and enol-N9H in the gas phase has been suggested.123,165 The double-proton transfer in adenine-thymine and guanine-cytosine base pairs at room temperature in gas phase and with the inclusion of environmental effects has been investigated.166,167 It was shown that double-proton transfer results in stable rare tautomeric forms only in guanine-cytosine base pairs.166,168,169
As known, there are four types of cyclobutane pyrimidine dimers: cis-syn, trans-syn, cis-anti and trans-anti dimers.170 This analysis only considers the cis-syn cyclobutane dimer, which is the most common type. There is no change in the orientation of the bases relative to the sugar-phosphate backbone as a result of cis-syn cyclobutane dimer formation.170 The mechanism of changes in tautomeric states is a radiation less deexitation of the quantum of UV energy.112,152 In result the hydrogen atom in hydrogen bond can assume new positions.112,146,150,152 It was shown what under cis-syn cyclobutane pyrimidine dimers formations the tautomer states of bases can change. A mechanism of changes in the tautomeric state of base pairs for the case when DNA molecule is UV-irradiated and cis-syn cyclobutane pyrimidine dimers are formed.122,146,150,152 As quantum-mechanical calculations show hydrogen atoms return in original position.166,168,169 Only one of new positions can be stable.
Assume that all processes of energy spreading are complete and that the UV-quantum is localized to one of the bases. This results in the excitation of electronic-vibration states.171 There are three channels for the relaxation of electronic-vibration states. First is fractional energy transfer to neighboring bases with radiation of some of the energy.172,173 This is the most probable process for the relaxation of a single electronic level,174 and it results in undamaged DNA. For a triplet level excitation, the most probable process for relaxation involves the transformation of energy to oscillations of the neighboring atoms.174 It is known172,173 that non radiative deexitation in DNA molecules occurs in a small volume of 3-5 bases pairs. This results in a strong "local heating-up", followed by initiation of normal oscillations of the bases. After several oscillations, taking ~10-14 - 10-12 sec,175,176 the vibratory system will reach equilibrium.
The oscillations of atoms will cause changes in the distances between the paired bases, in other words, in the lengths of H-bonds. For pairs of bases and in model systems, the first excited state results in a decrease in the height of potential energy barriers for proton transfer among H-bonds.177 For example, the barriers are lowered by a factor of approximately 1.5 for the Н-bond of a 7-azaindole dimer.177 Such change of proton potential occurs in ~ 10-16 sec and results in an increased probability that the hydrogen atom is in an excited state.178 Grebneva163 calculated the probabilities of a change in the lengths of Н-bonds (R) for distances of ±0.01 nm to±0.05 nm during several "instantaneous temperatures" of local heating-up.
Tolpygo and Grebneva175 described the shapes of the potential curves for the protons of all three H-bonds of Watson and Crick’s G: C base pair for several lengths of H-bonds. It turned out that a decrease in the length of a Н-bond by 0.02 nm from the equilibrium transforms the proton potential into a single-well. With an increase in Н-bond length, the second minimum becomes more and more obvious. The dynamics of the changes in the shape of proton potential for a wide spectrum of Н-bond lengths were calculated.179 The curves were designed with a system (water dimer, (Н2О)2) having H-bond parameters very similar to those of the H-bonds in base pairs.179 Semi-empirical potential function developed by us, has been used.180
The H-bonds that are formed between the DNA bases are characterized by a strong valence bond with one of the partner atoms in the Н-bond, and a weak bond with the other. When the Н-bond length (R) of a valence bond changes, the length (r) changes very little. But distance from the hydrogen to the second atom (R-r) varies considerably.181 A decrease in the R of a H-bond by 0.04-0.05nmis sufficient to place the atom of hydrogen almost in the center of Н-bond where it forms a strong bond. Therefore, when the Н-bond is extended, the hydrogen atom can assume new positions. If the atoms of hydrogen participating in the formation of Н-bonds are excited,178 their effective radius will increase by ~ 0.01 nm and there will be a higher probability that the hydrogen atom will participate in a Н-bond. Second, Danilova et al.177 found that potential energy barriers are reduced when the hydrogen’s are in an excited state. Third, some of the new proton positions can be stable, or even favorable from the point of view of the energy compared with other potential positions.166,168,169 The assumption of favorable conformations is possible, as the lifetime of a triplet state is ~10-9, 10-6, 10-12, 10-14 sec.174 The lifetime of the excited Н-bond is ~4 x 10-9 sec,182,183 and the characteristic periods of atomic oscillations are ~10-14 -10-12 sec.175 Therefore, there will be not more than several 10s of oscillations up to 100oscillations that influence the length of the H-bond. This results in a time of ~ 10-10 sec for any one conformation.
The proposed model is in a good agreement with the results obtained by other authors.185 Light-mediated proton transfer is required for the photo chromic transformation of organic compounds.176 Proton transitions in H-bonds occur in acids and bases, in crystals, proteins, molecular membranes, enzymes, and in other systems.176
Similar processes can occur upon dimer formation. The mechanism responsible for dimer formation and changes in tautomeric states is one and the same, namely, a radiation less deexitation of the quantum of UV energy. As a consequence, changes in the tautomeric state of bases can occur upon dimer formation. Hovorun178 has developed a model of semi open Meta stable states in DNA. Logically, the process of dimer formation proceeds through a semi open metastable state, and changes in the tautomeric conformation of bases follow the induction of metastable semi open states. The rare tautomeric states of thymine and adenine,152 which are represented in Figure 1, were obtained by assuming the occurrence of semi open metastable states.185
It was shown what under dimers formations the tautomer states of bases can change. A mechanism of changes in the tautomeric state of base pairs for the case when DNA molecule is UV-irradiated and thymine dimers are formed is the very mechanism when cytosine dimers are formed.112,152 The process of dimer formation proceeds through a semi open metastable state, and changes in the tautomeric conformation of bases follow the induction of metastable semi open states.112,152
In a canonical DNA molecule, conformational fluctuations of different DNA sites result in different metastable states .The exposure of bases from a double spiral to the solution is known as “opening” or “melting” of Watson and Crick’s pairs. In this case, the hydrogen bonds between DNA bases are broken.179 Metastable states developing with incomplete opening of the pair and with partial conservation of H-bonds and with the amine group free from H-bond are called semi open metastable states.180 There exist several models of semi open metastable states.180 In our opinion, the model developed by Hovorun177 is well-grounded; it explains a number of phenomena and is confirmed by experimental data.
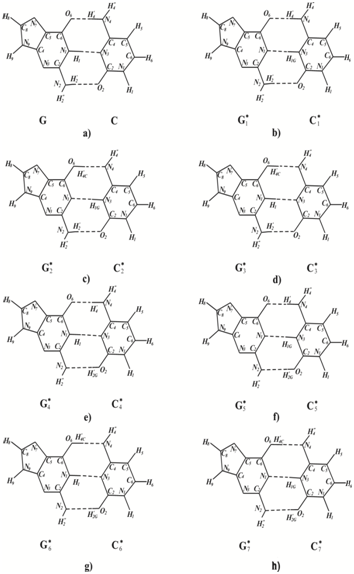
To find out what new tautomeric state can be formed at cytosine dimers formation, let us address to this structural-dynamical model of DNA semi open states by Hovorun.177 It is rather probable that the process of tautomeric state change goes in two stages. At the first stage, the metastable semi open DNA states are formed. At the second stage, the formed rare metastable tautomeric states transform into the stable ones. Let us study possible new tautomeric states of the Watson-Crick’s pair guanine-cytosine which may influence the character of pairing. According to Hovorun model,177 for guanine-cytosine pair there is several semi open states. It is well known that the lifetime of semi open states in DNA ~ 10-6 s.189 The processes for tautomeric state change are much less durable.112,153 Therefore, they may take place during the time of existence of semi open states. Figure 1 shows possible tautomeric states of bases of the G: C pair which could result from the processes described.
When cis-syn cyclobutane pyrimidine dimer is formed, the DNA strand containing this dimer becomes bent and the H-bonds become broken.181-185 That’s why in double-stranded DNA the mentioned rare tautomeric states are stable. DNA synthesis is rare quick even under the SOS synthesis, while it takes comparatively much time to change tautomeric states of bases because of the interaction with water molecules. Therefore these rare tautomeric forms will be also stable during DNA molecule synthesis when the DNA molecule is for a while in single-stranded form.112 The resulted were 7 rare tautomeric states of the cytosine and guanine Figure 1,146 that were capable of influencing the character of base pairing. Rare tautomeric forms of various types may occur with different probabilities. In Figure 1 the pairs of bases, which are in rare tautomeric forms, are enumerated in the order corresponding to the lowering of possible probabilities of their formation. Asterisk (*) means that the base is in a rare tautomeric form.
Cis-syn cyclobutane cytosine dimers cause mutations more frequent than cis-syn cyclobutane thymine dimers.27,28 The following analysis considers mechanisms for targeted base substitution mutations when the template DNA contains cis-syn cyclobutane cytosine dimers.
Error-prone and SOS replication of DNA containing Cis-Syn cyclobutane cytosine dimers
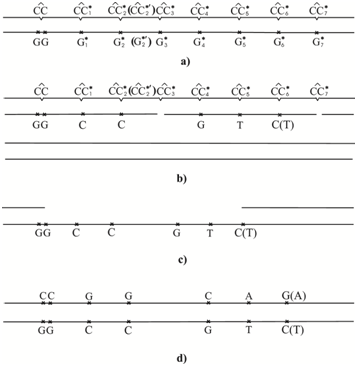
The polymerase-tautomer model for ultraviolet mutagenesis is based on that fact that during error-prone and SOS synthesis the induced DNA polymerase inserts canonical bases opposite the dimers; the inserted bases are capable of forming hydrogen bonds with bases in the template DNA. To determine which of the canonical bases will be inserted by the error-prone or SOS inducible DNA polymerase opposite cis-syn cyclobutane cytosine dimersFigure 2a consider the constraints on the formation of hydrogen bonds (H-bonds) between the bases of the template DNA and the inserted bases. DNA polymerase incorporates canonical bases capable of forming hydrogen bonds with dimerized bases in template DNA.112 First of all, H-bonds do not necessarily form. During error-prone or SOS synthesis of DNA containing cyclobutane pyrimidine dimers, nucleotide bases are inserted opposite the cyclobutane pyrimidine dimers without the removal of the dimer-containing sites. This only possible when the DNA polymerases, such as Pollδand Pollε of the eukaryotic cells or Poll III of the Escherichia coli are pressed on the DNA by the sliding clamp, obstructing the operation of exonuclease, or when the synthesis involves low-fidelity specialize DNA polymerases, such as Pollη and Pollζpolymerases of the yeast or Poll IV and Poll V polymerases of the Escherichia coli. Under these conditions, the strand of DNA containing no photo-dimers does not result in mutations Figure 2b.
The rare C1* cytosine tautomer Figure 1b does not form H-bonds with canonical guanine G because of the repulsion of hydrogen Н1 of guanine and Н1G of C1* cytosine atoms. Cytosine C1* is capable of forming two hydrogen bonds with adenine Figure 3a. The insertion of a canonical form of adenine opposite C1* produces G: C→А:T transition Figure 2. Canonical tautomeric forms of cytosine can be incorporated opposite the rare C1* Figure 1b cytosine tautomer Figure 3b. In this case, homologous G: C→C: G transverse will result Figure 5. The rare C1* tautomer cannot form H-bonds with canonical tautomer of thymine because of the repulsion of hydrogen atoms Н3of thymine and Н1G of C1* cytosine. So, rare C1* cytosine may produce G:C→А:T transition and homologous G:C→C:G transverse, but cannot cause G:C→T:A transverse or form error free pair G:C→G:C.
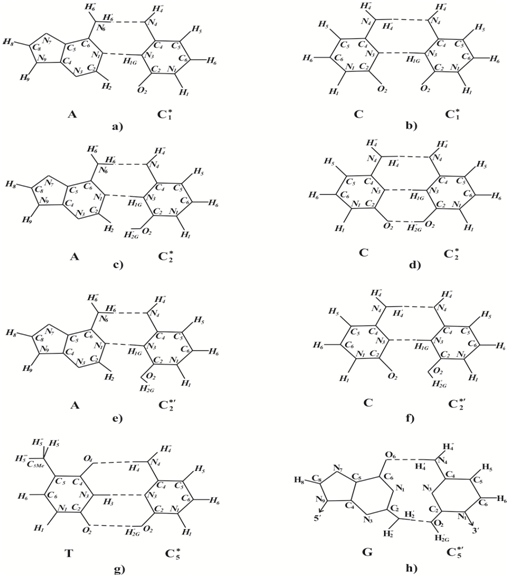
The rare C2* cytosine tautomer Figure 1c does not form hydrogen bonds with canonical guanine G because of the repulsion of hydrogen atoms Н1 of guanine and Н1G of C2* cytosine. But it can form two hydrogen bonds with canonical cytosine Figure 3d. The rare C2* cytosine tautomer is capable of forming two hydrogen bonds with adenine Figure 3c. Cytosine C2* cannot form H-bonds with canonical thymine because of the repulsion of hydrogen atoms Н3of thymine and Н1G of cytosine C2*. The insertion of a canonical form of cytosine opposite cytosine C2* produces homologous G: C→C: G transverse Figure 5, the insertion of a canonical form of adenine opposite cytosine C2* produces G:C→А:T transition Figure 2. If the Н¢2Ghydrogen atom in C2* cytosine turns as shown in Figure 3e, C2*¢ cytosine could form H-bond with adenine Figure 3e and cytosine Figure 3f. These pairings result in G: C→A: T transition Figure 2 or homologous G:C→C:G transverse Figure 5. So, the rare C2* and C2*¢ cytosine tautomer may produce G:C→А:T transition and homologous G:C→C:G transverse, but cannot cause G:C→T:A transverse or form error free pair G:C→G:C.
The rare C5* cytosine tautomer Figure 1f can’t form hydrogen bonds with canonical guanine G because of the repulsion of the Н¢2Ghydrogen of the cytosine C5* and hydrogen Н¢2 of guanine G and because of the repulsion of hydrogen atoms Н1 of guanine and Н1G of C5* cytosine. The rare C5* cytosine can’t form H-bonds with canonical cytosine because of the repulsion hydrogen Н¢4 of the cytosine C5* and hydrogen Н¢4 of the canonical cytosine. Cytosine C5* can’t form H-bonds with adenine because of the repulsion of the hydrogen Н¢4 of the cytosine C5* and hydrogen Н¢6 of the canonical adenine. But C5* can form three H-bonds with canonical thymine Figure 3g. The insertion of a canonical form of canonical thymine opposite cytosine C5* produces G:C→T:A transverse Figure 5. If the Н¢2G hydrogen atom in C5* cytosine turns as shown in Figure 3h, C5*¢cytosine could form H-bond with guanine Figure 3h, it may not result in mutations. So, the rare C5* cytosine tautomer may produce G:C→T:A transverse, but can’t cause G:C→А:T transition and homologous G:C→C:G transverse or form error free pair G:C→G:C.
The rare C6* cytosine tautomer Figure 1f can’t form hydrogen bonds with canonical guanine G because of the repulsion of the Н¢2Ghydrogen of the cytosine C6* and hydrogen Н¢2 of guanine G. The rare C6* cytosine tautomer Figure 1g can form two H-bonds with canonical cytosine Figure 4a, one H-bond with canonical adenine Figure 4c and two H-bonds with canonical thymine Figure 4b. Canonical tautomeric forms of cytosine, adenine or thymine can be incorporated opposite C6* cytosine. These insertions result in G:C→C:G homologous transverse Figure 5, G:C→A:T transition Figure 2 or G:C→T:A transverse Figure 5. If the H¢2G hydrogen atom in C6* cytosine turns as shown in Figure 4d, the resulting C6*¢ cytosine may form two H-bonds with canonical guanine Figure 4d. However, if the H¢¢4 hydrogen atom in C6* cytosine turns as shown in Figure 4e, C6*¢¢ cytosine could form three H-bonds with thymine Figure 4e. The insertion of a canonical tautomeric form of thymine opposite C6*¢¢cytosine can produce G:C→T:A transverse Figure 2. If the H¢2G hydrogen atom in C6*¢¢cytosine turns as shown in Figure 4f, the resulting C6*¢¢¢ cytosine may form three H-bonds with canonical guanine Figure 4f and it may not result in mutations. So, the rare C6*cytosine tautomer may produce G:C→A:T transition, G:C→C:G homologous transverse and G:C→T:A transverse, but can’t form error free pair G:C→G:C. The rare C6*¢¢ cytosine tautomer can produces G:C→T:A transverse, but can’t cause G:C→А:T transition and homologous G:C→C:G transverse or form error free pair G:C→G:C.
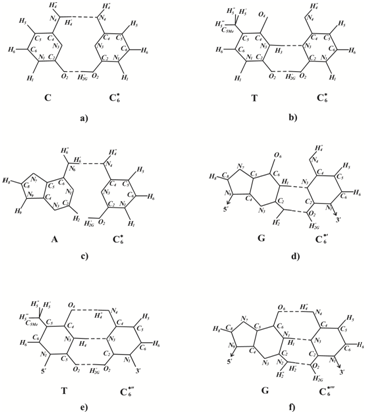
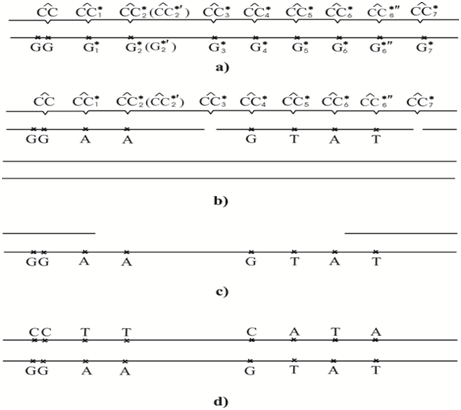
The rare C4* cytosine tautomer can pair with guanine; it may not result in mutations. The rare C3* and C7* cytosine tautomer do not form H-bonds with any canonical tautomer. So it is possible that DNA polymerase will not incorporate any bases opposite this rare tautomer and that DNA synthesis will result in a one-nucleotide gap. Figure 2 shows the most probable bases incorporated by the modified DNA-polymerase opposite Сi* cytosine. The CC3* and CC7* dimers will give gaps, and guanine will be likely incorporated opposite the CC4* dimer. Adenine will be incorporated opposite CC1*, CC2*, CC2*¢and CC6* dimers. Thymine will be incorporated opposite CC5* and CC6*¢¢dimers. It is known that the DNA strand having no dimers does not result in mutations Figure 2b. We shall consider that one-nucleotide gaps have been repaired, and the DNA site with dimers Figure 2b is repaired by the excision repair. Next, the DNA site having dimers is removed Figure 2c. The bases are inserted by an ordinary DNA polymerase in the excision gap Figure 2d. The complementary incorporation results in mutations: transitions G:C→A:T and transverse G:C→T:A.
Figure 5 shows another variant of events development. The CC3* and CC7* dimers will give gaps as be before. The CC4* dimer most likely will not result in mutation. The CC1*, CC2* and CC2*¢ dimers will cause the G:C→C:G homologous transverse. The CC5* dimer will induce G:C→Т:A transverse . The CC6* dimer can induce G:C→T:A transverse or G:C→C:G homologous transverse. These predictions need experimental verifications.
Let us interpret some experimental data from the viewpoint of the polymerase-tautomer model of UV mutagenesis. For example let us consider big collection of mutations caused by, cytosine photoproducts and cytosine-thymine photoproducts under hot spots UV mutagenesis investigation.27,28 We will investigate the UV mutational spectrum for supF (157G171C) variant having 90 mutations consisting of 68 G:C→A:T transitions, 14 homologous transversions G:C→C:G, two G:C→T:A transversions and six tandem mutations (when both bases of a photo dimer result in mutations).27,28 The CC photoproducts are sources of tandem mutations and all tandem mutations are G:C→A:T transitions. An interesting aspect of the mutations at 150 sites in the variant supF (157G171C) gene is that most of them were G: C→C: G transversions. This was quite different from almost all the other mutations which were G: C→A: T transitions. Homologous G: C→C: G transversions are not in accord with the “A-rule”. The polymerase-tautomer model explains these results in the following way.
Dipyrimidine photoproducts C99C100 and C103C104 give only transitions G: C→A:T. Dipyrimidine photoproduct C99C100 gives two transitions G:C→A:T opposite cytosine C99. So, they most likely correspond to the most probable cis-syn cyclobutane dimers C1*C. But it is quite probable that this is less possible cis-syn cyclobutane dimer C2*C or even cis-syn cyclobutane dimer C6*C. Moreover, dipyrimidine photoproduct C99C100 gives tandem mutation in the form of two transitions opposite cytosine C99 and cytosine C100 at a time. Then cis-syn cyclobutane dimer C1*C1* or, with all versions taken into account, cis-syn cyclobutane dimer Ci*Cj* (i=1, 2, 6; j=1, 2, 6) will correspond to the above cis-syn cyclobutane dimer, since only cytosine’s C1*, C2* and C6* can result in transitions G: C→A:T. Cis-syn cyclobutane dimer C103C104 gives four transitions G:C→A:T opposite cytosine C104. So, cis-syn cyclobutane dimer CCi* (i=1, 2, 6) will correspond to it. Dipyrimidine photoproduct C108C109 gives 11 transitions G: C→A:T and one transversions G:C→T:A opposite cytosine C108. The 11 transitions can be given only by cis-syn cyclobutane dimer Ci*C (i=1, 2, 6), while the transverse G:C→T:A can be only due to cis-syn cyclobutane dimers C5*C and C6*C, since only cytosine’s C5* and C6* can result in such a transverse. Dipyrimidine photoproduct C111T112 gives one homologous transverse G: C→C:G. It can be caused only by cis-syn cyclobutane dimer Ci*T, where i =1, 2, 6 since only cytosine’s C1*, C2* and C6* can result in homologous transversions G:C→C:G. Dipyrimidine photoproduct T112C113 gives one transition G:C→A:T, consequently, it is in correspondence with cis-syn cyclobutane dimer TCi* (i=1, 2, 6). Dipyrimidine photoproduct C124T125 gives one transition G:C→A:T, hence cis-syn cyclobutane dimer Ci*T (i=1, 2, 6) corresponds to it.
Dipyrimidine photoproduct C129T130 gives two transitions G: C→A:T opposite cytosine C129 and one transition A:T →G:C opposite thymine T130. In the first case, cis-syn cyclobutane dimer Ci*T (i =1, 2, 6) corresponds to it and in the second case - dimer CT1* since only T1* can result in transition A:T→G:C 112. Dipyrimidine photoproduct C133T134 gives four transitions G: C→A: T and one transverse G:C→T:A opposite cytosine C133 and one transverse A:T→T:A opposite thymine T134. In the first four cases, cis-syn cyclobutane dimer Ci*T (i =1, 2, 6) will correspond to it. In the fifth case - cis-syn cyclobutane dimer Ci*T (i=5, 6). In the sixth case, it will be cis-syn cyclobutane dimer CTi* (i=1, 5), since only thymines T1* and T5* may result in homologous transversions A: T→T: A [112]. Dipyrimidine photoproduct C139T140 gives one transition G: C→A: T opposite cytosine C139, consequently, there will be the correspondence with cis-syn cyclobutane dimer Ci*T (i=1, 2, 6).
Dipyrimidine photoproduct C150T151 gives two transitions G:C→A:T and twelve homologous transversions G:C→C:G opposite cytosine C150, consequently, cis-syn cyclobutane dimer Ci*T (i=1, 2, 6) will correspond to it. The larger number of transversions is, evidently, due to suppression of the enzymes responsible for the absence of pyrimidine - pyrimidine or purines - purines pairing, not due to some additional features of premutagenic lesion dimer. Dipyrimidine photoproduct T154C155 gives six transitions G: C→A: T opposite cytosine C155, so cis-syn cyclobutane dimer TCi* (i=1, 2, 6) will correspond to it. Dipyrimidine photoproduct C156C157 gives three transitions G: C→A:T opposite cytosine C157 and one tandem mutation consisting of two transitions G:C→A:T. In the first three cases there will be the correspondence with cis-syn cyclobutane dimer CCi* (i=1, 2, 6) and in the last case with cis-syn cyclobutane dimer Ci*Cj* (i=1, 2, 6; j=1, 2, 6). Dipyrimidine photoproduct C159C160 gives one transition G: C→A:T opposite cytosine C159, consequently, cis-syn cyclobutane dimer Ci*C (i=1, 2, 6) may correspond to it. Dipyrimidine photoproduct T162C163 gives six transitions G: C→A: T opposite cytosine C163, consequently, cis-syn cyclobutane dimer TCi* (i=1, 2, 6) will correspond to it. Dipyrimidine photoproduct C164T165 gives four transitions G: C→A: T opposite cytosine C164, consequently, cis-syn cyclobutane dimer Ci*T (i=1, 2, 6) will correspond to it. Dipyrimidine photoproduct C168C169 gives twelve transitions G: C→A: T, six ones due to cytosine C168 and the other six - cytosine C169, so in the first case, cis-syn cyclobutane dimer Ci*C (i=1, 2, 6) will correspond to it and in the latter case - cis-syn cyclobutane dimer CCi* (i=1, 2, 6). Dimer C172C173 gives three transitions G: C→A: T opposite cytosine C173, consequently, cis-syn cyclobutane dimer CCi* (i=1, 2, 6) will correspond to it. Dipyrimidine photoproduct C174C175 gives three transitions G: C→A:T opposite cytosine C174, so cis-syn cyclobutane dimer Ci*C (i=1, 2, 6) will correspond to it. Besides, it gives four tandem mutations in the form of transitions G: C→A: T, consequently, in this case, cis-syn cyclobutane dimer Ci*Cj* (i=1, 2, 6; j=1, 2, 6) will correspond to it.
So, the polymerase-tautomer model may explain all 90 substitution mutations. The most frequently arising transitions G: C→A:T and homologous transversions G:C→C:G may correspond the most probable rare C1* tautomeric forms of cytosine and two G:C→T:A transversions may correspond less probable rare C5* or C6* tautomeric form of cytosine. Predominance of homologous transversions G: C→C:G causing by dipyrimidine photoproduct C150T151 has naturally explanation since cytosine C1*, C2*, and C6* may cause homologous transversions G:C→C:G.
The polymerase-tautomer model for ultraviolet mutagenesis is described that is based on formation of rare tautomeric bases in cis-syn cyclobutane pyrimidine dimers. The model is based on that fact that during error-prone and SOS synthesis the induced DNA polymerase inserts canonical bases opposite the dimers; the inserted bases are capable of forming hydrogen bonds with bases in the template DNA. Error-prone and SOS replication of double-stranded DNA having cis-syn cyclobutane cytosine dimers, with one or both bases in a rare tautomeric conformation, results in targeted transitions and transversions. There are same types of potential mutagens damages in cis-syn cyclobutane cytosine dimers. They correspond to 7 fundamental types of rare tautomeric conformations of cytosine.
The structural analysis indicates that three types of cis-syn cyclobutane cytosine dimer containing a single tautomeric base (the cis-syn cyclobutane cytosine dimers CC1*, CC2*, CC2*¢) can cause G: C→A:T transition and G:C→C:G homologous transverse. Another two cis-syn cyclobutane cytosine dimers (the dimers CC5* and CC6*¢¢) can result only in G: C→Т: A transverse. The cis-syn cyclobutane cytosine dimer CC*6 can cause G: C→A:T transition, G:C→C:G homologous transverse or G:C→T:A transverse. The 90 substitution mutations are formed in the hot spots of UV mutagenesis27,28 are interpreted from point in view polymerase-tautomer model. The polymerase-tautomeric model may explain all 90 targeted base substitution mutations. The polymerase-tautomericmodel is able toexplain themechanisms formation of targeted base substitution mutations, targeted insertions, targeted deletions and targeted complex insertion, and hot and cold spots of UV-induced mutagenesis. The polymerase-tautomericmodel for bystander effects is able toexplain themechanisms formation for untargeted substitution mutations and targeted.
None.
Author declares that there is no conflict of interest.

©2016 Grebneva. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.