International Journal of
eISSN: 2381-1803


Research Article Volume 15 Issue 2
University College Cork, Ireland
Correspondence: Carina Harkin, University College Cork, Cork City, 24 Waterlane, Galway city, Co. Galway, Ireland
Received: March 30, 2022 | Published: April 12, 2022
Citation: Harkin C. Systematic review and meta-analysis of plant-derived antimicrobials in WHO priority pathogens. Int J Complement Alt Med. 2022;15(2):122-137. DOI: 10.15406/ijcam.2022.15.00600
Background: Antimicrobial resistance (AMR) threatens the effective prevention and treatment of a growing range of infections and requires immediate intersectoral action. A report into the global burden of AMR estimated that AMR contributed to 700,000 deaths in 2016 and is expected to reach 10 million deaths annually by 2050. Plant-derived antimicrobials (PDAms) have demonstrated the ability to combat multi-drug-resistant (MDR) pathogens and hope exists that they may help control AMR. This review investigated and recorded the antimicrobial activity (AMA) of whole plant ethanolic and methanolic extracts in WHO priority pathogens (WHO PPs) reported between 2000 until present. The review collated studies on PDAms tested on WHO PP laboratory isolates, determined efficacious PDAms, and grouped PDAms by WHO PP and antimicrobial susceptibility test (AST).
Methods: A comprehensive search of PubMed and Google databases using a combination of MeSH and free text terms was conducted from April 28th - May 8th 2020 of in-vitro studies determining the AMA of angiosperms in WHO PPs. Endpoints included minimum inhibitory concentration (MIC), zone of inhibition (ZOI) and half-maximal inhibitory concentration (IC50). This systematic review and meta-analysis was reported according to PRISMA guidelines. A modified STROBE-AMS checklist was applied to included studies. Limitations are that a double screening selection process is generally Scholar recommended.
Results: Data were pooled and computed in a meta-analysis. Thirty-four studies on PDAm-WHO priority specific data were identified. Effective PDAms were grouped by WHO PP and AST. PDAm activity was demonstrated in 50% of WHO PPs. Piper betle L. demonstrated the greatest AMA in six WHO PPs (50%). Methicillin-resistant Staphylococcus aureus (MRSA) was the most commonly studied WHO PP (69.6%), returning forty-three PDAms. No studies were returned on clarithromycin-resistant Helicobacter pylori, fluoroquinolone-resistant Campylobacter spp. fluoroquinolone-resistant Salmonellae, penicillin-non-susceptible Streptococcus pneumoniae, ampicillin-resistant Haemophilus influenzae and fluoroquinolone-resistant Shigella spp.
Conclusion: This synthesis of the available studies and identification of the quality data may help to inform and prioritise future research on PDAms in WHO PPs.
AMA, antimicrobial activity; AMR, antimicrobial resistance; AMRIC, the antimicrobial resistance and infection control division; AMU, antimicrobial use; AB, antibiotics; AR, antibiotic resistance; AST, antimicrobial susceptibility testing; CPE, carbapenemase-producing enterobacterales; CLSI, clinical and laboratory standards institute; CAM, complementary and alternative medicine; EUCAST, european committee on antimicrobial susceptibility testing; E. coli, ESBL-E. Escherichia coli; ESBL-E, extended-spectrum β-lactamase-producing enterobacterales; GPB, gram positive bacteria; GNB, gram negative bacteria; IC50, half maximal inhibitory concentration; HCAIs, healthcare associated infections; HPSC, health protection surveillance centre; MRSA, methicillin-resistant staphylococcus aureus; MIC, minimum inhibitory concentration; MBC, minimum bactericidal concentration; MDR, multidrug resistant; MDRO, multidrug resistant organisms; PDAms, plant-derived antimicrobials; QS, quorum sensing; R&D, research and development; RMAs, resistance-modifying agents; STROBE-AMS, STROBE for antimicrobial stewardship; SSIs, surgical site infections; VRE, vancomycin-resistant Enterococci; WHO, world health organisation; WHO PPs, WHO priority pathogens; ZOI, zone of inhibition
Antimicrobial Resistance (AMR) occurs when microorganisms, including bacteria, fungi, viruses and parasites, adapt to become resistant to their counterpart antimicrobial drugs. Antimicrobial-resistant microbes are found in people, animals, food, water, soil and air, and can spread from person to person or person to animal. The spread is magnified by poor infection control, sanitation and food handling.1 Overprescribing antimicrobials is the main driver of AMR.2 A report by Jim O’Neill report entitled ‘The Review on Antimicrobial Resistance’ found that AMR contributed to 700 000 deaths in 2016 and is predicted to reach 10 million deaths annually by 2050. The foreword of this report admitted that this figure did not account for the secondary effects of AMR such as the risks imposed by surgery.3 Additionally, former UK Chief Medical Officer (CMO) Sally Davies highlighted the magnitude of underreporting of AMR deaths and called for AMR to be put on relevant death certificates.4 A Royal Institute of International Affairs review on AMR progress, estimated a cost to the global economy of US$100 trillion by 2050.5
Antibiotic resistance
Antibiotic Resistance (AR) is a subset of AMR and applies to bacteria becoming resistant to antibiotics.6 In 2017 the WHO published a WHO priority pathogens (WHO PPs) list of AR bacteria requiring urgent research and development (R&D) for new antibiotics.7 The WHO PPs are a catalogue of twelve families of bacteria that are categorised into three priority levels. The WHO PP list was developed in collaboration with the Division of Infectious Diseases Tübingen University whereby a multicriteria decision analysis was used to prioritise these AR bacteria. These criteria included infections with the highest mortality, length of hospital stay required, how frequently resistance is encountered once community spread occurs, the ease of zoonotic or human-to-human transmission, preventability through hygiene practices and or vaccines, therapeutics, and whether new AB were already in the R&D channel.8 Table 1 describes the WHO Priority Pathogens. (Appendix 1 & Appendix 2).
Priority 1 |
Acinetobacter baumannii (carbapenem-resistant A. baumannii) |
Priority 2 |
Enterococcus faecium (vancomycin-resistant E. faecium) [vancomycin-resistant Enterococci (VRE)] |
Priority 3 Medium |
Streptococcus pneumoniae (penicillin-non-susceptible S. pneumoniae) |
Table 1 WHO Priority Pathogens
Healthcare-associated infections
The Society of Hospital Medicine (SHM) describes the WHO PPs as the principle nosocomial pathogens.9 Healthcare-associated infections (HCAIs) lead to an increase in morbidity, mortality and healthcare costs and are a public health emergency. The WHO estimates hundreds of millions of patients are affected by HCAIs annually, yet admits the difficulty in gathering reliable data means the true burden is unknown.10 The WHO rank HCAIs as the fifth greatest threat to global health and one of the top ten causes of hospital deaths worldwide.11 AMR is a key driver of HCAIs and increases associated morbidity and mortality.12 The leading cause of HCAIs is a group of common multidrug-resistant (MDR) bacteria known as ESKAPE pathogens (E. faecium, S. aureus, K. pneumoniae, A. baumannii, P. aeruginosa and Enterobacter spp.).13 The ESKAPE pathogens are extended-spectrum beta-lactamase (ESBL)-producing, the latter four are Gram-negative bacteria (GNB) and all are WHO PPs.14 As the principal ESBL-Es, E. coli and K. pneumoniae have been chosen for the purpose of this review.15
A 2017 Point Prevalence Survey of HCAIs and antimicrobial use (AMU) in European Acute Care Hospitals identified the following WHO PPs, methicillin-resistant Staphylococcus aureus (MRSA), vancomycin-resistant Enterococci (VRE), extended-spectrum β lactamase (ESBLs) and carbapenem-resistant or carbapenemase-producing Enterobacterales (CPE), as the pathogens associated with higher healthcare costs, increased length-of-stay and higher mortality.16 The Antimicrobial Resistance and Infection Control (AMRIC) division section of Ireland’s Health Protection Surveillance Centre (HPSC) state the pathogens associated with AMR in Ireland are CPE, VRE, E. coli, K. pneumoniae, P. aeruginosa and N. gonorrhoeae.17
Considering again that the O’Neill report estimate of 10 million deaths annually did not account for the secondary effects of AMR, it is important to contemplate the potential additional global burden. Surgical site infections (SSIs) for example, are one of the most common HCAIs and are associated with increased morbidity and mortality.18 77% of surgical patient deaths are related to a SSI.19 Globally, 300 million people undergo operations annually with 31% resulting in a SSI.20 Given that between 39% and 51% of SSI pathogens are AR, the figure of 10 million deaths annually is potentially substantially underestimated.21
Sepsis is another secondary effect of AMR, with AMR posing a major challenge in sepsis treatment. Paradoxically, antimicrobial stewardship programmes and sepsis are at odds, with delayed, incomplete and ineffective treatment driving AMR and increasing sepsis risk.22 Current estimates are that 20% of all global deaths are sepsis-related. A review into the global burden of sepsis states that AMR may be a major driver of community and hospital-acquired sepsis.23 A recent study estimated that in 2017 there were 48.9 million cases of sepsis worldwide and 11 million deaths. Previous global sepsis burden estimates were significantly lower at 30 million cases leading to 6 million deaths.24 The WHO global priority pathogens list is one of the WHO projects aimed to prevent, diagnose and treat sepsis.25 A study looking at the contribution of sepsis to mortality in patients with sepsis from two independent cohorts, the Kaiser Permanente Northern California (KPNC) and the Healthcare Cost and Utilization Project (HCUP) Nationwide Inpatient Sample (NIS), found that sepsis contributed to 1 in every 2 to 3 hospital cohort deaths.26 The reviewers of a 2015 report, ‘Just Say Sepsis’, by the National Confidential Enquiry into Patient Outcome and Death (NCEPOD) found that in cases where sepsis was not mentioned as a cause of death on the death certificate that it should have been in 48/59 (81.4%) of cases.27
Plant-derived antimicrobials
The O’Neill report describes the potential for both vaccines and alternatives to lower antimicrobial use (AMU). These alternatives include phage therapy, lysins, antibodies, probiotics, and peptides and immune stimulation; described specifically as boosting the patient's natural immune system.3 Herbal medicine is a complementary and alternative medicine (CAM). Herbal Medicine boosts the patient's natural immune system. Herbal immunomodulators have been shown to stimulate the innate and adaptive defence mechanisms of the host to attune the activity of the immune system, thus decrease the inflammatory response.28
Pharmacognosy describes the study of plants as drug sources. It is estimated that 25% of modern drugs are directly or indirectly plant-derived. Additionally, more than one third of all FDA-approved new molecular entities (NMEs) are of natural origin.29,30 120 plant secondary metabolite phytochemicals have been isolated and identified, with 80% showing a positive correlation between modern therapeutic and traditional use.31,32 In herbal pharmacology, the sixteen main phytochemical groups are alkaloids, anthocyanins, anthraquinones, cardiac glycosides, coumarins, cyanogenic glycosides, flavonoids, glucosinolates, phenols, saponins and tannins.33 Phytochemical bioactivity is of particular clinical value as plant strategies to deal with microorganisms adapt over time hence are generally considered not to confer resistance.34,35 Herbal resistance in clinical bacterial isolates has been demonstrated however and needs to be considered. E. coli herbal resistant plasmid for example, has been shown to replicate and be expressed in human urinary tract S. aureus.36,37
Ethnopharmacology offers a wide array of plant-derived antimicrobials (PDAms) and increasing numbers of pharmaceutical companies are focusing on botanical drug development.38,39 Evidence suggests PDAms display broad-spectrum activity against both Gram-positive bacteria (GPB) and GNB, and can attenuate bacterial virulence.40,41 PDAms have exhibited activity against virulence factors including quorum sensing (QS), biofilms, motility, toxins, pigments, enzymes and bacterial surfactants.42,43 Antibiotic discovery is not keeping up with the evolution of resistance and urgency exists to find novel antimicrobial agents. Plant secondary metabolite phytochemicals and whole plant extracts have demonstrated their potential as either stand-alone antibacterials or as potentiators of conventional antibacterial agents. As yet however, the therapeutic application of PDAms remains to be clinically proven.
The relevance of in-vitro antimicrobial susceptibility testing (AST) in ascertaining the antimicrobial activity (AMA) of plants cannot be overemphasised. Due to use of various non-standardised approaches including inoculum preparation method, inoculum size, growth medium, atmosphere, temperature, incubation duration and end-point determination however, comparisons between results are difficult. ASTs themselves have certain advantages and disadvantages. Aiming to ensure an accurate experimental approach is used that can allow a comparison of results, accepted and approved standards are published by the Clinical and Laboratory Standards Institute (CLSI) and the European Committee on Antimicrobial Susceptibility Testing (EUCAST), two global organisations that establish interpretive criteria for in-vitro susceptibility data.44. Table 2 provides a summary of the pros and cons of in-vitro ASTs used in PDAms.
AST |
Pros |
Cons |
Diffusion Methods |
|
|
Agar disk diffusion |
Widely used to determine the AMA of plants.Provides zones of inhibition (ZOI) (Balouiri M. et al. 2016; p. 74). Standards provided by the CLSI and EUCAST are qualitative; can be quantitative according to CLSI categories of susceptible, intermediate and resistant (Jorgensen and Ferraro, 2009; p.1751). ZOI can be compared with that of different antibiotic standards in an antibiogram and described as susceptible, intermediate or resistant (Balouiri M. et al. 2016; p. 72). |
Not all fastidious bacteria can be tested accurately however standardisation allows testing of certain fastidious bacterial pathogens (Balouiri M. et al., 2016; p. 72). Impossibility to measure amount of test compound diffused into agar medium. Not appropriate to determine MIC; MIC values are approximated using stored algorithms (Balouiri M. et al. 2016; p. 72). |
Agar well diffusion |
Widely used to determine the AMA of plants. Provides ZOI (Balouiri M. et al. 2016; p. 74). |
|
Thin-layer chromatography (TLC)–bioautography |
Used to separate complex mixtures into bioactive compounds (Balouiri M. et al. 2016; p. 74). |
Bioautography is not a quantitative measure of antimicrobial activity |
Dilution methods |
|
|
Broth microdilution method |
Quantitative measurement. The gold standard for minimum inhibitory concentration (MIC) determination (Lallemand E.A. et al., 2016; p 1). Provides the most consistent results, allowing determination of relative species sensitivities and relative AMA of each medium (King T. et al., 2008; p. 1423). Can be used to calculate the half maximal inhibitory concentration (IC50), the concentration of a drug required to inhibit 50% of a microorganism in vitro. IC50 is a more accurately definable measure than the MIC and standard error is easily calculable (Soothill J.S. et al.,1992; p. 137). |
|
Broth macrodilution method |
MIC determination. |
Tedious, manual and high risk of error (Balouiri M. et al. 2016; p. 75). |
Agar dilution method |
Agar dilution is preferred over broth dilution in determining MIC when the compound/ extract being tested is coloured and masks microbial growth detection in the liquid medium (Balouiri M. et al. 2016; p. 76). Possible to estimate concentration of test compound in agar medium therefore the most commonly used (Balouiri M. et al. 2016; p. 76). Standards provided by CLSI and EUCAST (Balouiri M. et al. 2016; p. 75). |
|
Table 2 Summary of in-vitro ASTs used in PDAms: Pros and Cons
A group of international experts came together through a joint initiative by the European Centre for Disease Prevention and Control (ECDC) and the Centers for Disease Control and Prevention (CDC) to create a standardized international terminology with which to describe acquired resistance profiles in Staphylococcus aureus, Enterococcus spp., Enterobacteriaceae (other than Salmonella and ), Pseudomonas aeruginosa and Acinetobacter spp., all bacteria often responsible for healthcare-associated infections and prone to multidrug resistance. Clinical Laboratory Standards Institute (CLSI), the European Committee on Antimicrobial Susceptibility Testing (EUCAST) and the United States Food and Drug Administration (FDA).45
A variety of herbal preparation methods exists also. The European Medicine Agency and the European Directorate for the Quality of Medicines and HealthCare state the most commonly used solvents are ethanol, methanol and water. Ethanol and methanol are frequently used in herbal extraction techniques such as maceration, percolation and soxhlet extraction as their polarity maximises phytochemical extraction and complies with their European Pharmacopoeia monographs.46,47
Previous systematic reviews on the cidal activity of PDAms in MDR-bacteria exist, however these review the phytochemical classes of plant-derived resistance-modifying agents (RMAs) to potentiate other antibacterial agents. None of these reviews are on WHO PP specific susceptible strains of bacteria.48-50 As no reviews on PDAMs and WHO specific PPs exist, this systematic review and meta-analysis is a worthwhile study and may contribute to the body of existing research.
The aim of this review was to collate information on PDAms tested on WHO PP bacterial isolates and to determine the most efficacious PDAms.
Study objectives were to conduct a systematic review and meta-analysis of the data in order to answer the research question.
PDAms have demonstrated efficacious AMA in WHO PPs.
In applying the feasible, interesting, novel, ethical, and relevant (FINER) Cochrane standards and the PICO technique requiring consideration of patient population, intervention (exposure), comparison (control) and outcome, the research question is which PDAms have demonstrated AMA in WHO PPs and which of these has demonstrated the greatest AMA in each WHO PP.51,52
Study design
This systematic review and meta-analysis was reported according to the Preferred Reporting Items for Systematic Review and Meta-Analysis (PRISMA) framework.53.
Study Selection
Table 3 displays the inclusion/exclusion criteria that set the boundaries for the review.
Table 3 Inclusion/exclusion criteria
Search strategy
Table 4 displays the search strategies for bibliographic databases used in conducting the systematic review.
|
Concept 1 |
Concept 2 |
Concept 3 |
Concept 4 |
Key concepts |
Antimicrobial Resistance |
Botanical Medicine |
Anti-Bacterial |
Antimicrobial susceptibility tests |
Free text terms / natural language terms |
Antibiotic resistance, |
Herbal Medicine,
|
Antibacterial,
|
Minimum Inhibitory Concentration (MIC), |
Controlled vocabulary terms / Subject terms |
Drug Resistance, Microbial (NCBI [Internet] Drug Resistance, Microbial, 1963) |
Angiosperms. |
Anti-Bacterial Agents (NCBI [Internet] Drug Resistance, Microbial, 1963) |
Microbial Sensitivity Test. |
Table 4 Search strategies for bibliographic databases98
Information sources
The MeSH database was consulted to identify concepts and choose appropriate terms and combinations of free-text and controlled-vocabulary terms. Prior to establishing these search criteria, a scoping exercise was used to ensure that the search terms ‘drug resistance, microbial’ returned individual WHO PPs and the search term ‘angiosperms’ returned the most diverse group of PDAms being flowering plants (mosses and ferns were excluded). An online literature search of PubMed /Google Scholar bibliographic databases was conducted from April 28th –May 8th 2020 using a combination of MeSH and free text terms. The exact search terms used in both PubMed and Google Scholar were; Drug Resistance, Microbial, Angiosperms, Antimicrobial Tests; returning 1115 and 17100 results respectively. It was intended that 1000 results only be downloaded from Google Scholar. In the event where “Drug Resistance, Microbial” alone did not return a particular WHO PP, the specific pathogen was searched in addition although this tactic did not produce any additional returns.
Data management
Google scholar was set at 10 results per page. When activity was flagged as suspicious a virtual private network (VPN) service provider was used to change the IP address and the CAPTCHA I am not a robot checked. The Zotero plug-in was used to export citations. Only 98 page results were accessible, totalling 980 returns in total rather than the planned 1000. All PubMed results were imported into Endnote, 200 at a time, using “send to citation manager” providing 1115 results in total. Endnote was used to store, manage, merge and remove duplicates.
First level single screening
A first level screening of titles and abstracts was conducted and inclusion/exclusion criteria applied.54
Second level screening
Full text articles were critically appraised using a specifically devised modified STROBE for antimicrobial stewardship (STROBE-AMS) checklist for PDAm ASTs. The modified STROBE was used to assess the internal and external validity of the study, reduce the risk of bias and assure methodological quality and quality of evidence.55,56 See Figure 2: modified STROBE-AMS checklist.
Data collection processes
Data was extracted. Included studies were grouped by WHO PP and AST result. Eight texts were received through Research Gate PDF requests with one only meeting the modified Strobe-AMS criteria.
Data items
The PICO items for which data were sought included WHO PP, whole PDAm methanol or ethanol extracts only and PDAm activity in WHO PP demonstrated.
Outcomes and prioritisation
The primary outcome was to retrieve a sufficient number of studies determining the AMA of PDAms in WHO PPs. The secondary outcome was to determine the most efficacious PDAm in each WHO PP. Full data is included in the appendices.
Data synthesis and analysis
Data were pooled by pathogen, plant, AST and computed in a meta-analysis.
Figure 1 shows the study flow diagram based on Moher D et al., PRISMA 2009 Flow Diagram.
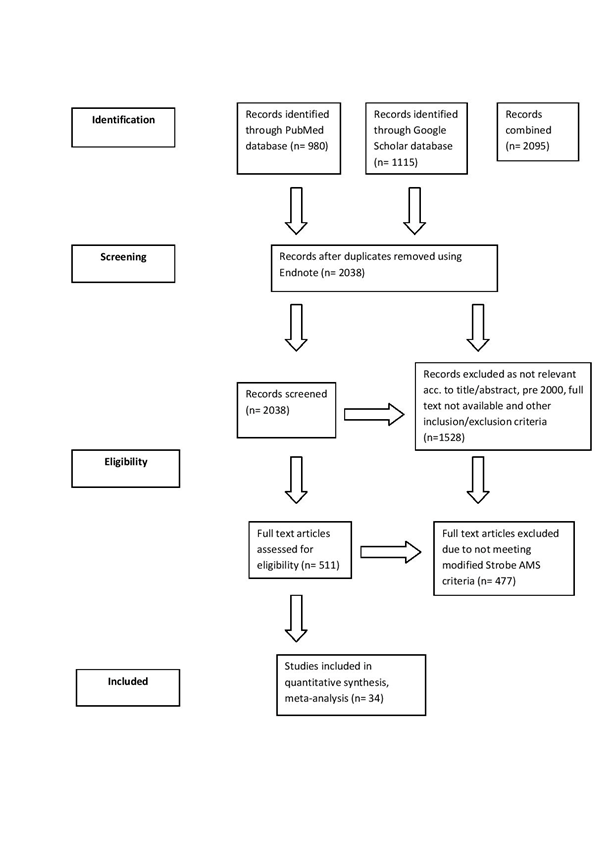
Figure 1 Study flow diagram based on Moher D et al.,53 PRISMA 2009 Flow Diagram.
34 studies on PDAm-WHO priority specific data were identified. PDAm activity was demonstrated in 50% of WHO PPs. Piper betle L. demonstrated the greatest AMA in 7 out of the 13 WHO PPs (53.9%). S. aureus was the most commonly studied pathogen (69.6%), returning 43 PDAm species. Results data are tabulated below.
Priority 1
PDAm activity was demonstrated in all three priority 1 pathogens; carbapenem-resistant A. baumannii and P. aeruginosa, and in ESBL‐producing and carbapenem-resistant E. coli and K. pneumoniae Enterobacter spp. 4 out of 34 (11.8%) studies returned confirmed PDAm activity in carbapenem-resistant A. baumannii. 7 PDAms demonstrated AMA. Piper betel L. demonstrated both the lowest MIC and greatest ZOI.57-60 Table 5 indicates that Piper betle L. demonstrated the greatest AMA against carbapenem resistant A. baumannii with the lowest MIC of 406 μg/ml.57 Terminalia bellirica demonstrated a similar MIC of 500 μg/ml.58 Table 6 indicates that Martynia annua L. IC50 was 256 μg/ml. Table 7 indicates Piper betle L. demonstrated the highest ZOI of 24mm.57
Plant part and solvent used |
MIC μg/ml |
Piper betle L. (leaf ethanol) |
406 (mean of 5 MDR A.baumannii strains) (Valle DL Jr. et al., 2016; p.9) |
Terminalia bellirica (fruit methanol) |
500 (Dharmaratne MPJ. et al., 2018; p. 8) |
Piper betle L. (leaf methanol) |
526.4 (mean of 5 MDR A.baumannii strains) (Valle DL Jr. et al., 2016; p.9) |
Rosmarinus officinalis (leaf ethanol): |
1000 (Assis FV. et al., 2018; p. 1667) |
Plectranthus barbatus (leaf ethanol): |
2000 (mean of 2 A.baumannii strains) |
Ocimun basilicum (leaf ethanol): |
2000 (Assis FV. et al., 2018; p. 1667) |
Mentha spp. (leaf ethanol): |
2000 (Assis FV. et al., 2018; p. 1667) |
Table 5 MIC Results: Acinetobacter baumannii (carbapenem-resistant)
Plant part and solvent used |
IC50 μg/ml |
Martynia annua L. (leaf ethanol): |
256 (Khan MF. et al., 2018; p. 6) |
Table 6 IC50 Results: Acinetobacter baumannii (carbapenem-resistant)
Plant part and solvent used |
ZOI mm |
Piper betle L. (leaf ethanol) |
24 (mean of 5 MDR A.baumannii strains) (Valle DL Jr. et al., 2016; p.7) |
Terminalia bellirica (fruit methanol) |
17 (mean of 2 A.baumannii strains) |
Table 7 ZOI Results: Acinetobacter baumannii (carbapenem-resistant)
4 out of 34 (11.8%) studies returned confirmed PDAm activity in carbapenem-resistant P. aeruginosa. 7 PDAms demonstrated AMA. Piper betel L. demonstrated both the lowest MIC and greatest ZOI.57-59, 61 Table 8 indicates that Piper betle L. demonstrated the greatest AMA in the average of 3 strains, exhibiting the lowest MIC of 260 μg/ml.57 Plectranthus barbatus and Terminalia bellirica demonstrated equal MICs of 500 μg/ml.58,59 Table 9 indicates that Piper betle L. demonstrated the largest ZOI of 21.33mm followed by Terminalia bellirica of 17.55 mm and then Nigella sativa L. of 13mm.57, 58, 61
Plant part and solvent used |
MIC μg/ml |
Piper betle L. (leaf ethanol) |
260 (Valle DL Jr. et al., 2016; p.9) |
Terminalia bellirica (fruit methanol) |
500 (Dharmaratne MPJ. et al., 2018; p. 8) |
Plectranthus barbatus (leaf ethanol) |
500 (Assis FV. et al., 2018; p. 1667) |
Mentha spp. (leaf ethanol) |
2000 (Assis FV. et al., 2018; p. 1667) |
Ocimun basilicum (leaf ethanol) |
>2000 (Assis FV. et al., 2018; p. 1667) |
Rosmarinus officinalis (leaf ethanol) |
>2000 (Assis FV. et al., 2018; p. 1667) |
Table 8 MIC Results: Pseudomonas aeruginosa (carbapenem-resistant)
Plant part and solvent used |
ZOI mm |
Piper betle L. (leaf ethanol) |
21.33mm (Valle DL Jr. et al., 2016; p.7) |
Terminalia bellirica (fruit methanol) |
17.55 (Dharmaratne MPJ. et al., 2018; p. 7) |
Nigella sativa L. (leaf methanol) |
13mm P < 0.001 (Salman MT. et al., 2009; p.99) |
Table 9 ZOI Results: Pseudomonas aeruginosa (carbapenem-resistant)
13 out of 34 (38%) studies confirmed PDAm activity in Enterobacterales species. Of these Enterobacterales species, 6 out of 34 studies were returned for E. coli and 7 out of 34 studies were returned for K. pneumoniae. 4 out of 34 (11.8%) studies returned confirmed PDAm activity in ESBL-producing E. coli. 7 PDAms demonstrating AMA. Nauclea latifolia demonstrated the lowest MIC and Piper betel L. demonstrated the greatest ZOI.57,58, 62, 63 Table 10 indicates that Nauclea latifolia demonstrated the lowest MIC of 32 μg/ml. Table 11 indicates that Piper betle L. demonstrated the greatest ZOI of 20mm. 2 out of 34 (5.9%) studies confirmed PDAm activity in carbapenem-resistant E. coli. 2 PDAms demonstrated AMA. Croton campestris demonstrated the lowest MIC.64, 65 Table 12 indicates that Croton campestris demonstrated the lowest MIC of 512 μg/ml.
Plant part and solvent used |
MIC μg/ml |
Nauclea latifolia (stem bark methanol) |
32 (Tekwu EM.,et al, 2012; p. 269) |
Albizia gummifera (stem bark and leaves methanol) |
128 (Tekwu EM. et al, 2012; p. 269) |
Ficus exasperate (leaf methanol) |
128 (Tekwu EM. et al., 2012; p. 269) |
Ricinodendron heudelotii (stem bark methanol) |
312 (Tekwu EM. et al., 2012; p. 269) |
Piper betle L. (leaf ethanol) |
312 (Valle DL Jr. et al., 2016; p.9) |
Terminalia bellirica (fruit methanol) |
>500 (Dharmaratne MPJ. et al., 2018; p. 8) |
Vitex doniana (bark methanol) |
7142 (Ouattara A. et al., 2013; p 99) |
Table 10 MIC Results: Escherichia coli (ESBL‐producing)
Plant part and solvent used |
ZOI mm |
Piper betle L. (leaf ethanol) |
20 (Valle DL Jr. et al., 2016; p.7) |
Albizia gummifera (stem bark and leaves methanol) |
16–20 (Tekwu EM., et al, 2012; p. 268) |
Ficus exasperate (leaf methanol) |
16–20 (Tekwu EM., et al, 2012; p. 268) |
Nauclea latifolia (stem bark methanol) |
16–20 (Tekwu EM., et al, 2012; p. 268) |
Ricinodendron heudelotii (stem bark methanol) |
11–15 (Tekwu EM., et al, 2012; p. 268) |
Vitex doniana (bark methanol) |
13 (Ouattara A., et al., 2013; p 97) |
Terminalia bellirica (fruit methanol) |
12 (Dharmaratne MPJ. et al., 2018; p. 7) |
Table 11 ZOI Results: Escherichia coli (ESBL‐producing)
Plant part and solvent used |
MIC μg/ml |
Croton campestris (leaf methanol) |
512 (Matias EF. et al., 2011; p.307) |
Murraya paniculata (leaf ethanol) |
1024 (Menezes IR. et al., 2015; p. 5) |
Table 12 MIC Results: Escherichia coli (carbapenem-resistant)
4 out of 34 (11.8%) studies returned confirmed PDAm activity in ESBL-producing K. pneumoniae. 4 PDAms demonstrated AMA. Piper betel L. demonstrated both the lowest MIC and greatest ZOI.57,58,60,63 Table 13 indicates that Piper betle L. demonstrated the lowest MIC of 625μg/ml. Table 14 indicates that Martynia annua L. demonstrated an IC50 of 256 μg/ml. Table 15 indicates that Piper betle L. demonstrated the highest ZOI of 20mm. 3 out of 34 (8.8%) studies returned confirmed PDAm activity in carbapenem-resistant K. pneumoniae. 8 PDAms demonstrated AMA. Piper betel L. demonstrated both the lowest MIC and greatest ZOI.57,59,66 Table 16 indicates that Piper betle L. demonstrated the lowest MIC of 390μg/ml. Table 17 indicates that Alstonia scholaris demonstrated the lowest mean IC50 of 4060 μg/ml. Table 18 indicates that Piper betle L. demonstrated the largest ZOI of 23mm.
Plant part and solvent used |
MIC μg/ml |
Piper betle L. (leaf ethanol) |
625 (Valle DL Jr. et al., 2016; p.9) |
Terminalia bellirica (fruit methanol) |
>5000 (Dharmaratne MPJ. et al., 2018; p. 8) |
Vitex doniana (bark methanol) |
71420 (Ouattara A. et al., 2013; p 99) |
Table 13 MIC Results: Enterobacterales: Klebsiella pneumoniae (ESBL‐producing)
Plant part and solvent used |
IC50 μg/ml |
Martynia annua L. (leaf ethanol): |
256 (Khan MF. et al., 2018; p. 6) |
Table 14 IC50 Results: Klebsiella pneumonie (ESBL‐producing)
Plant part and solvent used |
ZOI mm |
Piper betle L. (leaf ethanol) |
20 (Valle DL Jr. et al., 2016; p.7) |
Terminalia bellirica (fruit methanol) |
12 (Dharmaratne MPJ. et al., 2018; p. 7) |
Vitex doniana (bark methanol) |
11 (Ouattara A. et al., 2013; p 97) |
Table 15 ZOI Results: Klebsiella pneumoniae (ESBL‐producing)
Plant part and solvent used |
MIC μg/ml |
Piper betle L. (leaf ethanol) |
390 (Valle DL Jr. et al., 2016; p.9) |
Rosmarinus officinalis (leaf ethanol) |
500 (Assis FV. et al., 2018; p. 1667) |
Plectranthus barbatus (leaf ethanol) |
>2000 (Assis FV. et al., 2018; p. 1667) |
Ocimun basilicum (leaf ethanol) |
>2000 (Assis FV. et al., 2018; p. 1667) |
Mentha sp. (leaf ethanol) |
>2000 (Assis FV. et al., 2018; p. 1667) |
Table 16 MIC Results: Klebsiella pneumoniae (carbapenem-resistant)
Plant part and solvent used |
Mean IC50 values 8 hr (μg/ml) |
Alstonia scholaris (stem ethanol) |
4060 μg/ml (Bonvicini F. et al., 2014; p. 3) |
Tinospora cordifolia (stem ethanol) |
7070 μg/ml (Bonvicini F. et al., 2014; p. 3) |
Crataeva nurvala (stem ethanol) |
>12500 μg/ml (Bonvicini F. et al., 2014; p. 3) |
Table 17 IC50 Results: Klebsiella pneumoniae (carbapenem-resistant)
Plant part and solvent used |
ZOI mm |
Piper betle L. (leaf ethanol) |
23 (Valle DL Jr. et al., 2016; p.7) |
Table 18 ZOI Results: Klebsiella pneumoniae (carbapenem-resistant)
Priority 2
PDAm activity was demonstrated in 3 out of 6 priority 2 pathogens; vancomycin-resistant E. faecium, methicillin-resistant and vancomycin-intermediate S. aureus and cephalosporin-resistant and fluoroquinolone-resistant N. gonorrhoeae. 3 out of 34 (8.8%) studies confirmed PDAm activity in vancomycin-resistant E. faecium. 5 PDAms demonstrated AMA. Piper betel L. demonstrated both the lowest MIC and greatest ZOI.57, 60,67 Table 19 indicates that Piper betle L. demonstrated the lowest MIC of 64.67μg/ml. Table 20 indicates that Adiantum capillus, Artemisia absinthium and Zanthoxylum armatum demonstrated the same IC50 of 256 μg/ml. Table 21 indicates that Piper betle L. demonstrated the largest ZOI of 28.3 mm.
Plant part and solvent used |
MIC μg/ml |
Piper betle L. (leaf ethanol) |
64.67 (Valle DL Jr. et al., 2016; p.9) |
Table 19 MIC Results: Enterococcus faecium (vancomycin-resistant)
Plant part and solvent used |
IC50 μg/ml |
Adiantum capillus (whole plant ethanol) |
256 (Khan MF. et al., 2018; p. 6) |
Artemisia absinthium (aerial parts ethanol) |
256 (Khan MF. et al., 2018; p. 6) |
Zanthoxylum armatum (fruit ethanol) |
256 (Khan MF. et al., 2018; p. 6) |
Table 20 IC50 Results: Enterococcus faecium (vancomycin-resistant)
Plant part and solvent used |
ZOI mm |
Piper betle L. (leaf ethanol) |
28.3 (Valle DL Jr. et al., 2016; p.7) |
Cinnamomum porrectum (stem bark methanol) |
7.5 ± 0.7071(Buru AS. Et al., 2014; p. 3) |
Table 21 ZOI Results: Enterococcus faecium (vancomycin-resistant)
23 out of 34 (66.7%) studies confirmed PDAm activity in MRSA. 43 plant species demonstrated AMA.57, 58, 60, 62, 65-83 3 of these studies reported results in mean IC50 values.60,66,82 Tecoma stans demonstrated the lowest MIC, Castanea sativa demonstrated the lowest IC50 and Piper betel L. demonstrated the greatest ZOI. Table 22 indicates that Tecoma stans demonstrated the lowest MIC of 7.81 μg/ml. Table 23 indicates that Castanea sativa demonstrated the lowest IC50 of 16 μg/ml. Table 24 indicates that Piper betle L. demonstrated the largest ZOI of 32.7mm. Table 24 also indicates that Thuja orientalis and Psidium guajava had similar ZOIs of >29mm. 1 out of 34 (2.9%) of studies confirmed PDAm activity in vancomycin-intermediate S. aureus. 9 PDAms demonstrated AMA. Punica granatum and Anogeissus acuminate equally demonstrated the lowest MIC and the greatest ZOI.84 Table 25 indicates that Anogeissus acuminate and Punica granatum demonstrated the lowest MIC of 290μg/ml. Table 26 indicates that Anogeissus acuminate also demonstrated the largest ZOI of 27 mm followed closely by Punica granatum at 26mm.
Plant part and solvent used |
MIC μg/ml |
Tecoma stans (leaf ethanol) |
7.81 (Bakr RO. et al., 2019; p. 5) |
Nauclea latifolia (stem bark methanol) |
64 (Tekwu EM. et al, 2012; p. 269) |
Zingiber officinale (rhizome ethanol) |
68.45 (Karuppiah and Rajaram, 2012; p. 600) |
Allium sativum (cloves ethanol) |
78.90 (Karuppiah and Rajaram, 2012; p. 600) |
Quercus infectoria (gall ethanol) |
80 (Wan Nor Amilah WA. et al., 2014; p. 5) |
Terminalia fagifolia (stem bark methanol) |
100 (de Araujo AR. et al., 2015; p. 6) |
Piper betle L. (leaf ethanol) |
167.14 (Valle DL Jr. et al., 2016; p.9) |
Punica granatum (seeds ethanol) |
250 (all 16 MRSA strains) (Machado TB. et al. p. 282) |
Tabebuia avellanedae (wood ethanol) |
250 (all 16 MRSA strains) (Machado TB. et al. p. 282) |
Terminalia bellirica (fruit ethanol) |
250 (Dharmaratne MPJ. et al., 2018; p. 8) |
Albizia gummifera (stem bark and leaves methanol) |
256 (Tekwu EM. et al, 2012; p. 269) |
Hygrophila auriculata (root methanol) |
500 (Savitha and Yoganant, 2016; p. 198) |
Jasminum sambac (leaf ethanol) |
500 (Al-Hussaini and Mahasneh, 2009; p. 3430-31) |
Jasminum sambac (flower ethanol) |
500 (Al-Hussaini and Mahasneh, 2009; p. 3430-31) |
Lavandula angustifolia (flower) |
500 (Al-Hussaini and Mahasneh, 2009; p. 3430-31) |
Laurus nobilis (fruit ethanol) |
500 (Al-Hussaini and Mahasneh, 2009; p. 3430-31) |
Laurus nobilis (leaf ethanol) |
500 (Al-Hussaini and Mahasneh, 2009; p. 3430-31) |
Laurus nobilis (bark ethanol) |
500 (Al-Hussaini and Mahasneh, 2009; p. 3430-31) |
Populus nigra (leaf) |
500 (Al-Hussaini and Mahasneh, 2009; p. 3430-31) |
Rosmarinus officinalis (leaf ethanol) |
500 (Al-Hussaini and Mahasneh, 2009; p. 3430-31) |
Tecoma capensis (flower ethanol) |
500 (Al-Hussaini and Mahasneh, 2009; p. 3430-31) |
Ficus exasperate (leaf and stem bark methanol) |
512 (Tekwu EM. et al, 2012; p. 269) |
Ricinodendron heudelotii (stem methanol) |
512 (Tekwu EM. et al, 2012; p. 269) |
Chuquiraga straminea (flowers methanol) |
533.3 μg/ml [average 3 strains] (Mendiondo ME. et al., 2011; p. 966) |
Cinnamomum porrectum (stem bark methanol) |
625.00 ±0 (Buru AS. Et al., 2014; p. 7) |
Cinnamomum iners (stem bark methanol) |
625.00 ±0 (Buru AS. Et al., 2014; p. 7) |
Sonchus oleraceus (ariel ethanol) |
750 (Al-Hussaini and Mahasneh. 2009; p. 3430-31) |
Laurus nobilis (flower ethanol) |
1000 (Al-Hussaini and Mahasneh. 2009; p. 3430-31) |
Rosmarinus officinalis (flower ethanol) |
1200 (Al-Hussaini and Mahasneh, 2009; p. 3430-31) |
Populus alba (leaf) |
1500 (Al-Hussaini and Mahasneh, 2009; p. 3430-31) |
Tecoma capensis (leaf ethanol) |
1500 (Al-Hussaini and Mahasneh, 2009; p. 3430-31) |
Aloe barbadensis (leaf ethanol) |
1560 (Dahiya and Purkayastha 2010;p. 447) |
Azadirachta indica (leaf ethanol) |
1560 (Dahiya and Purkayastha 2010;p.447) |
Ocimum sanctum (leaf ethanol) |
1560 (and Purkayastha 2010;p.447) |
Scrophularia striata (whole plant methanol) |
3100 (Zangeneh M. et al., 2017; p. 42) |
Origanum vulgare (leaf ethanol) |
3120 (Dahiya and Purkayastha 2010;p. 447) |
Rosmarinus officinalis (flower ethanol) |
3120 (Dahiya and Purkayastha 2010;p. 447) |
Thymus vulgaris (leaf ethanol) |
3120 (Dahiya and Purkayasth 2010;p. 447) |
Rafflesia kerrii Meijer (perigone lobe ethanol) |
6000 [average 5 MRSA isolates] (Hirunpetcharat C. et al., 2018; p. 43) |
Piper sarmentosum (leaf methanol) |
50000 (Fernandez L. et al., 2012; p. 109) |
Table 22 MIC Results: Staphylococcus aureus (methicillin-resistant)
Plant part and solvent used |
IC50 μg/ml |
Castanea sativa (leaf methanol) |
16 (Quave CL et al., 2015; p.13) |
Adiantum capillus (aerial parts ethanol) |
256 (Khan MF. et al., 2018; p. 6) |
Martynia annua L. (fruit ethanol) |
256 (Khan MF. et al., 2018; p. 6) |
Swertia chirata (whole plant ethanol) |
256 (Khan MF. et al., 2018; p. 6) |
Zanthoxylum armatum (fruit ethanol) |
256 (Khan MF. et al., 2018; p. 6) |
Tinospora cordifolia (fruit ethanol) |
12280 [10.81 – 13.95] (Bonvicini F. et al., 2014; p. 3) |
Alstonia scholaris (fruit ethanol) |
8080 (7.25 – 9.00) (Bonvicini F. et al., 2014; p. 3) |
Crataeva nurvala (fruit ethanol) |
6150 (4.75– 7.85) (Bonvicini F. et al., 2014; p. 3) |
Table 23 IC50 Results: Staphylococcus aureus (methicillin-resistant)
Plant part and solvent used |
ZOI mm |
Piper betle L. (leaf ethanol) |
32.7 (Valle DL, Jr. et al., 2016; p.7) |
Thuja orientalis (leaf ethanol) |
29.80±0.71 (Chakraborty S. et al., 2018; p.354) |
Psidium guajava (leaf ethanol) |
29.69±0.78(Chakraborty S. et al., 2018; p.354) |
Hygrophila auriculata (root methanol) |
22 (Savitha and Yoganant, 2016; p. 198) |
Terminalia bellirica (fruit ethanol) |
20.5 (Dharmaratne MPJ. et al., 2018; p. 7) |
Scrophularia striata (whole plant methanol) |
19 (Zangeneh M. et al., 2017; p. 42) |
Tecoma stans (leaf ethanol) |
18.3±0.25 (Bakr RO. et al., 2019; p. 5) |
Ocimum sanctum (leaf ethanol) |
18.10±0.10 (Dahiya and Purkayasth 2010;p. 446) |
Laurus nobilis (bark ethanol) |
18±0.7 (Al-Hussaini R and Mahasneh AM. 2009; p. 3429) |
Aloe barbadensis (leaf ethanol) |
17.90±0.35 (Dahiya and Purkayasth 2010;p. 446) |
Azadirachta indica (leaf ethanol) |
16.96±0.10 (Dahiya and Purkayasth 2010;p. 446) |
Jasminum sambac (flower ethanol) |
16±0.7 (Al-Hussaini and Mahasneh, 2009; p. 3429) |
Laurus nobilis (leaf ethanol) |
16±0.7 (Al-Hussaini R and Mahasneh AM. 2009; p. 3429) |
Lavandula angustifolia (flower) |
15±0.7 (Al-Hussaini and Mahasneh, 2009; p. 3429) |
Origanum vulgare (leaf ethanol) |
14.90±0.05 (Dahiya and Purkayasth 2010;p. 446) |
Allium sativum (cloves ethanol) |
14.55±0.20 (Karuppiah and Rajaram 2012; p. 599) |
Rosmarinus officinalis (flowers ethanol) |
14.00±0.90mm (Dahiya and Purkayastha 2010;p. 447) |
Thymus vulgaris (leaf ethanol) |
13.76±0.20 (Dahiya and Purkayasth 2010;p. 446) |
Zingiber officinale (rhizome ethanol) |
13.55±0.20 (Karuppiah and Rajaram 2012; p. 599) |
Quercus infectoria (gall ethanol) |
13.33 ± 0.33 P value 0.52 (Wan Nor Amilah WA. et al., 2014; p. 4) |
Rosmarinus officinalis (flower ethanol) |
13±0.7 (Al-Hussaini R and Mahasneh AM. 2009; p. 3429) |
Tecoma capensis (flower ethanol) |
12.5±0.7 (Al-Hussaini R and Mahasneh AM. 2009; p. 3429) |
Rosmarinus officinalis (leaf ethanol) |
12±2.1 (Al-Hussaini and Mahasneh,2009; p. 3429) |
Laurus nobilis (fruit ethanol) |
12±2.1 (Al-Hussaini R and Mahasneh AM. 2009; p. 3429) |
Albizia gummifera (stem bark and leaves methanol): |
11–15 mm (Tekwu EM., et al, 2012; p. 268) |
Ficus exasperate (leaf and stem bark methanol) |
11–15 (Tekwu EM., et al, 2012; p. 268) |
Nauclea latifolia (stem bark methanol) |
11–15 (Tekwu EM., et al, 2012; p. 268) |
Ricinodendron heudelotii (stem methanol) |
11–15 (Tekwu EM., et al, 2012; p. 268) |
Laurus nobilis (flower ethanol) |
11±0.0 (Al-Hussaini R and Mahasneh AM. 2009; p. 3429) |
Rafflesia kerrii Meijer (perigone lobe ethanol) |
11.1 (Hirunpetcharat C. et al., 2018; p. 43) |
Cinnamomum porrectum (stem bark methanol) |
10.5 ± 0.7071 (Buru AS. Et al., 2014; p. 7) |
Populus nigra (leaf) |
10±1.4 Al-Hussaini and Mahasneh, 2009; p. 3429) |
Sonchus oleraceus (ariel ethanol) |
10±1.4 (Al-Hussaini R and Mahasneh AM. 2009; p. 3429) |
Piper sarmentosum (leaf methanol) |
10.0±0.0 (p=0.0018) (Fernandez L. et al., 2012;p. 109) |
Cinnamomum iners (stem bark methanol) |
9.5 ± 0.7071 (Buru AS. Et al., 2014; p. 7) |
Tecoma capensis (leaf ethanol) |
8.0±1.4 (Al-Hussaini and Mahasneh, 2009; p. 3429) |
Populus alba (leaf) |
8.0±1.4 (Al-Hussaini and Mahasneh, 2009; p. 3429) |
Table 24 ZOI Results: Staphylococcus aureus (methicillin-resistant)
Plant part and solvent used |
MIC μg/ml |
Anogeissus acuminate (leaf bark methanol) |
290 (Mishra MP. et al., 2017; p.90) |
Punica granatum (leaf, bark, fruits methanol) |
290 (Mishra MP. et al., 2017; p.90) |
Soymida febrifuga (leaf, bark methanol) |
670 (Mishra MP. et al., 2017; p.90) |
Terminalia chebula (leaf methanol): MIC μg/ml, ZOI 23 mm |
1510 (Mishra MP. et al., 2017; p.90) |
Azadirachta indica (leaf methanol) |
3410 (Mishra MP. et al., 2017; p.90) |
Bauhinia variegate (leaf methanol) |
3410 (Mishra MP. et al., 2017; p.90) |
Tribulus terrestris (leaf, bark methanol): MIC μg/ml, ZOI 21 mm |
3410 (Mishra MP. et al., 2017; p.90) |
Boerhaavia diffusa (leaf, root methanol) |
4270 (Mishra MP. et al., 2017; p.90) |
Tinospora cordifolia (leaf, fruits methanol): |
4270 (Mishra MP. et al., 2017; p.90) |
Table 25 MIC Results: Staphylococcus aureus (vancomycin-intermediate)
Plant part and solvent used |
ZOI mm |
Anogeissus acuminate (leaf bark methanol) |
27 (Mishra MP. et al., 2017; p.89) |
Punica granatum (leaf, bark, fruits methanol) |
26 (Mishra MP. et al., 2017; p.89) |
Soymida febrifuga (leaf, bark methanol) |
25 (Mishra MP. et al., 2017; p.89) |
Terminalia chebula (leaf methanol) |
23 (Mishra MP. et al., 2017; p.89) |
Bauhinia variegate (leaf methanol) |
21 (Mishra MP. et al., 2017; p.89) |
Tribulus terrestris (leaf, bark methanol) |
21 (Mishra MP. et al., 2017; p.89) |
Azadirachta indica (leaf methanol) |
20 (Mishra MP. et al., 2017; p.89) |
Tinospora cordifolia (leaf, fruits methanol) |
19 (Mishra MP. et al., 2017; p.89) |
Boerhaavia diffusa (leaf, root methanol) |
17 (Mishra MP. et al., 2017; p.89) |
Table 26 ZOI Results: Staphylococcus aureus (vancomycin-intermediate)
No studies were returned on priority 2 clarithromycin-resistant H. pylori, fluoroquinolone-resistant Campylobacter spp. and fluoroquinolone-resistant Salmonellae. 1 out of 34 (2.9%) studies confirmed PDAm activity in fluoroquinolone-resistant N. gonorrhoea. 2 PDAms demonstrated AMA. Tagetes erecta L and Myrteola nummularia demonstrated equal ZOI of 27 mm.85 Table 27 indicates that Tagetes erecta L and Myrteola nummularia demonstrated equal ZOI of 27 mm.
Plant part and solvent used |
ZOI mm |
Tagetes erecta L. (flower methanol) |
> 20 mm (Ruddock PS. et al. 2011; p.84) |
Myrteola nummularia (aerial parts methanol) |
> 20 mm (Ruddock PS. et al. 2011; p.84) |
Table 27 ZOI Results: Neisseria gonorrhoeae (fluoroquinolone-resistant)
1 out of 34 (2.9%) studies confirmed PDAm activity in both fluoroquinolone-resistant and cephalosporin-resistant N. gonorrhoeae. The fluoroquinolone-resistant and cephalosporin less sensitive WHO K and L strains were equally highly sensitive to all 15 plant extracts tested. The AMA of the plant extracts against N. gonorrhoeae was detected by measuring the ZOI. The percentage inhibition was then determined by considering inhibition by penicillin (0.5 IU) and ciprofloxacin (1 g) to be 100% for antibacterial activity and comparing the extracts ZOIs with these standards.86 Table 28 indicates that all N. gonorrhoeae (cephalosporin-resistant, fluoroquinolone-resistant) were highly sensitive to all 15 PDAms with the percentage inhibition of the ZOI being ≥100%.
Plant part and solvent used |
ZOI a No inhibition. b 1–50% inhibition. c 51–99% inhibition. d ≥100% inhibition |
Adhatoda vasica (roots methanol) |
highly sensitive ≥100% inhibition (Shokeen, Bala and Tandon, 2009; p. 88-9) |
Albizzia lebbeck (stem bark methanol) |
highly sensitive ≥100% inhibition (Shokeen, Bala and Tandon, 2009; p. 88-9) |
Alangium salviifolium (seeds methanol) |
highly sensitive ≥100% inhibition (Shokeen, Bala and Tandon, 2009; p. 88-9) |
Anethum sowa (fruits methanol) |
highly sensitive ≥100% inhibition (Shokeen, Bala and Tandon, 2009; p. 88-9) |
Azadirachta indica (leaf methanol) |
highly sensitive ≥100% inhibition (Shokeen, Bala and Tandon, 2009; p. 88-9) |
Cedrela toona (leaf methanol) |
highly sensitive ≥100% inhibition (Shokeen, Bala and Tandon, 2009; p. 88-9) |
Drynaria peregrine (stem bark methanol) |
highly sensitive ≥100% inhibition (Shokeen, Bala and Tandon, 2009; p. 88-9) |
Eugenia camaldulensis (leaf methanol) |
highly sensitive ≥100% inhibition (Shokeen, Bala and Tandon, 2009; p. 88-9) |
Euphorbia hirta (entire plant methanol) |
highly sensitive ≥100% inhibition (Shokeen, Bala and Tandon, 2009; p. 88-9) |
Michelia champaca (stem bark methanol) |
highly sensitive ≥100% inhibition (Shokeen, Bala and Tandon, 2009; p. 88-9) |
Phyllanthus fraternus (entire plant methanol) |
highly sensitive ≥100% inhibition (Shokeen, Bala and Tandon, 2009; p. 88-9) |
Plumeria rubra (leaves, stem and stem bark [3 separate preparations] methanol) |
highly sensitive ≥100% inhibition (Shokeen, Bala and Tandon, 2009; p. 88-9) |
Ricinus communis (stem and roots [2 separate preparations] methanol) |
highly sensitive ≥100% inhibition (Shokeen, Bala and Tandon, 2009; p. 88-9) |
Salvadora persica (leaves and stem [2 separate preparations] methanol) |
highly sensitive ≥100% inhibition (Shokeen, Bala and Tandon, 2009; p. 88-9) |
Urginea indica (bulbs methanol) |
highly sensitive ≥100% inhibition (Shokeen, Bala and Tandon, 2009; p. 88-9) |
Table 28 ZOI Results: Neisseria gonorrhoeae (cephalosporin-resistant, fluoroquinolone-resistant
Priority 3
No studies were returned on penicillin-non-susceptible S. pneumoniae, ampicillin-resistant H. influenza and fluoroquinolone-resistant Shigella spp. Table 29
Priority 1 |
% of total studies |
Nu. PDAms |
PDAm with Lowest MIC |
MIC |
PDAm with Largest ZOI |
ZOI |
A. baumannii (carbapenem-resistant) |
11.8 |
7 |
Piper betle L. |
406 |
Piper betle L. |
24 |
Pseudomonas aeruginosa (carbapenem-resistant) |
11.8 |
5 |
Piper betle L. |
260 |
Piper betle L. |
21.3 |
Enterobacterales |
|
|
|
|
|
|
E. coli TOTAL |
17.7 |
|
|
|
|
|
E. coli |
11.8 |
7 |
Nauclea latifolia |
32 |
Piper betle L. |
20 |
E. coli |
5.9 |
2 |
Croton campestris |
512 |
Not results returned |
N/A |
K. pneumoniae TOTAL |
20.6 |
|
|
|
|
|
K. pneumoniae |
11.8 |
4 |
Piper betle L. |
625 |
Piper betle L. |
20 |
K. pneumoniae |
8.8 |
8 |
Piper betle L. |
390 |
Piper betle L. |
23 |
Priority 2 |
|
|
|
|
|
|
E. faecium |
8.8 |
5 |
Piper betle L. |
64.67 |
Piper betle L. |
28.3 |
S. aureus TOTAL |
69.6 |
|
|
|
|
|
S. aureus (methicillin-resistant) |
66.7 |
43 |
Tecoma stans |
80 |
Piper betle L. |
32.7 |
S. aureus (vancomycin-intermediate) |
2.9 |
9 |
Anogeissus acuminate and Punica granatum |
290 |
Anogeissus acuminate |
27 |
H. pylori (clarithromycin-resistant) |
0 |
|
|
|
|
|
Campylobacter spp. (fluoroquinolone-resistant) |
0 |
|
|
|
|
|
Salmonellae (fluoroquinolone-resistant) |
0 |
|
|
|
|
|
N. gonorrhoeae |
5.8 |
|
|
|
|
|
N. gonorrhoeae (fluoroquinolone-resistant) |
2.9 |
2 |
No results returned |
N/A |
Tagetes erecta and |
>20 |
N. gonorrhoeae (cephalosporin-resistant, fluoroquinolone-resistant) |
2.9 |
15 |
No results returned |
|
15 Indian plant |
≥100% |
Priority 3 |
|
|
|
|
|
|
S. pneumoniae (non penicillin-non-susceptible) |
0 |
|
|
|
|
|
H. influenza (ampicillin-resistant) |
0 |
|
|
|
|
|
Shigella spp. (fluoroquinolone-resistant) |
0 |
|
|
|
|
|
Table 29 Main Summary Findings
Promising results were found for Priority 1 Critical Pathogens, including carbapenem-resistant A. baumannii and P. aeruginosa, Carbapenemase-producing Enterobacterales (CPE) and extended-spectrum beta-lactamase producing Enterobacterales (ESBL-E). Furthermore, results were found for vancomycin-resistant E. feacium, a vancomycin-resistant Enterococcus (VRE), another principal nosocomial pathogen. As zero studies were returned on clarithromycin-resistant H. pylori, fluoroquinolone-resistant Campylobacter spp., fluoroquinolone-resistant Salmonellae, penicillin-non-susceptible S. pneumoniae, ampicillin-resistant H. influenza and fluoroquinolone-resistant Shigella spp. as this review analysed over 2000 studies, it can be safely postulated that further research on these pathogens is urgently required.
Moreover and frustratingly, many studies identified PDAm activity in MDR bacterial isolates yet could not be included in the analysis as they were not WHO PP specific. Such studies either confirmed sensitivity to the antibiotics mentioned in the WHO PP list or did not test these specific antibiotics. For example, three studies referred to MDR A. baumannii and identified multiple antibiotic-resistant isolates however did not specify carbapenem resistance.69, 78, 84 Four studies referred to MDR P. aeruginosa, identifying many antibiotic-resistant isolates using the multiple antibiotic resistance (MAR) index that tests ciprofloxacin, vancomycin and methicillin, however did not specify carbapenem-resistance.62,72,78,84 Eight studies referred to MDR E. coli., identifying multiple antibiotic-resistant isolates however did not specify carbapenem-resistance or ESBL-producing.59,72,75, 80, 84, 87-89 Five studies referred to MDR K. pneumoniae as per MAR index, again identifying multiple antibiotic-resistant isolates however failed to specify carbapenem-resistance or ESBL-production.62,72,75, 84, 87 Three studies confirmed PDAm activity in drug resistant Salmonellae, yet strains were not identified as fluoroquinolone resistant. One study identified the Salmonellae strain as chloramphenicol-resistant however fluoroquinolones were not tested.62 Another study identified Salmonellae strains as chloramphenicol-resistant however these were susceptible to the fluoroquinolone ciprofloxacin.90 All Salmonellae strains in one study were identified as MDR however were also identified as susceptible to ciprofloxacin.88 Whilst no studies on penicillin-non-susceptible S. pneumoniae were returned, one study confirmed PDAm activity in drug resistant S. pneumoniae however the strain identified was the macrolide erythromycin and penicillin was not tested.90
Piper betel L. was the most frequently mentioned plant antimicrobial. Piper betel L belongs to the Piperaceae family. The leaves are used in traditional medicine for the treatment of a variety of ailments.91 Notably it is one ingredient of 'betel quid' or 'paan’ and is commonly chewed in Asia as a mild stimulant.92 In addition to its antibacterial properties, studies have shown that Piper betel L. is anticaries, antifungal, antioxidant, anti-inflammatory, antiprotozal, antiulcer, chemoprotective, gastroprotective, hepatoprotective, immunomodulatory and larvicidal.93 Whilst the phytochemical and pharmacological profiling of Piper betel L is well established, and biotechnological interventions have aimed to improve the cash value of this botanical, as yet, no biotechnological interventions have progressed toward the development of a medical product.94, 95
It is vitally important to note that this review is on laboratory studies (in-vitro) only not by choice, but as this is the only evidence available. Botanical drug development is only emerging. Recently, an antibiotic-producing strain of Streptomyces sp. TM32 was isolated from of the medicinal plant Curcuma longa L.96 Whilst the US Food and Drug Administration (FDA) has established a clear regulatory pathway for botanical drugs, to date, only two drugs have been approved.97 It is the hope that the information obtained in these pre-clinical phase trials may one day work towards clinical testing in humans, yet until these laboratory studies progress through to clinical trials, the scientific community is a long way off ameliorating microbial resistance to drugs in humans using PDAms.
Further limitations are that a double screening selection process is generally recommended to enable the uncertainty to be quantified with more confidence and to eliminate bias. Whilst CLSI/ EUCAST have standardised AST procedures there can be no absolute surety that comparisons are meaningful. This research would benefit by expanding search criteria to include plant bioactive compounds and alternative solvents.
Risk of bias, quality of evidence assignment
Some PDAms demonstrated a MIC >2000 μg/ml hence were resistant but as results were so limited, these were included. Whilst a checklist was used for unbiased decision making, a risk of bias still exists.
AMR is one of the greatest threats to global health. It is promising that PDAms have demonstrated AMA in all WHO Priority 1 Critical and the majority of Priority 2 High pathogens. It is particularly promising that the principal nosocomial infections, the ESKAPE pathogens, MRSA, CPE, ESBL-E and VRE are represented, as addressing these presents the greatest opportunity to reduce HCAIs. Research on the Priority 2 High pathogens clarithromycin-resistant H. Pylori, fluoroquinolone-resistant Campylobacter spp., fluoroquinolone-resistant Salmonellae and in all Priority 3 Medium pathogens is urgently required. Given the public health emergency posed by the WHO PPs, future PDAm studies should prioritise these specific resistant bacterial isolates. This review has highlighted that PDAms have demonstrated in-vitro anti-bacterial activity in WHO PPs and contributed to the body of data. As it stands, the therapeutic application of PDAms remains to be clinically proven. Given the global burden of AR and the urgent need for novel antibacterial agents, and considering that preclinical plant antimicrobial studies have demonstrated promise perhaps it is pertinent that some allocation of the funding mentioned in the O’Neill report be directed toward finding novel antimicrobial agents from plants, with an aim to progress botanical drug development to tackle drug-resistant infections globally. Most certainly, more research on PDAms in WHO specific PPs is urgently required.
Ethics approval and consent to participate.
The SPH Ethics Department of UCC approved the research project on the 28th April 2020.
Not applicable.
The authors confirm that the data supporting the findings of this study are available within the article appendices.
The author declares that there is no conflict of interest.
No funding was allocated to this project.
Carina Harkin contributed to the design and implementation of the research protocol and to the writing of the manuscript.
The author would like to thank Dr. Brendan Palmer and Dr Éilis O'Reilly from the Dept. of Epidemiology and Public Health University College Cork (UCC) for their direction and feedback.

©2022 Harkin. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.