International Journal of
eISSN: 2381-1803


Research Article Volume 7 Issue 6
1Departamento de Ciências Biológicas, Universidade Estadual de Santa Cruz, Brazil
2Departamento Ciências Agrárias, Universidade Estadual de Santa Cruz, Brazil
3Faculdade de Farmácia, Universidade Federal do Rio Grande do Sul, Brazil
4Comissão Executiva do Plano da Lavoura Cacaueira, Brazil
5Departamento de Biofísica, Universidade Federal do Rio Grande do Sul, Brazil
Correspondence: C Pungartnik, Departamento de Ciências Biológicas, Universidade Estadual de Santa Cruz, Rod. Ilhéus Itabuna, Km 16, Ilheu, BA, Brazil, Tel +55 73 36805336
Received: February 09, 2017 | Published: July 11, 2017
Citation: Santos RX, Oliveira D, Sodré GA, Gosmann G, Brendel M et al. (2017) Fermented Theobroma Cacao Pod Husk Extract: Genotoxicity in Mammalian Cells, Yeast and Bacteria. Int J Complement Alt Med 7(6): 00243. DOI: 10.15406/ijcam.2017.07.00243
Theobroma cacao L. is a plant of economic importance to the region south of Bahia. Of culture cocoa is generated a large amount of waste from the rind of the fruit, which is underused. New technologies were encouraged to use and reduction of this waste so that cocoa can be fully used. We therefore evaluated the cocoa pod husks (CHE) revealed that levels of transition metals such a zinc (Zn), iron (Fe), manganese (Mn), and copper (Cu). We also evaluate sensitivity and genotoxicity/mutagenicity of CHE in trials established test (comet assay, tests in Saccharomyces cerevisiae and Salmonella/microsome test). It was observed that yeast cells harboring STAT fet3Δ, aft1Δ and mutant alleles were sensitive to CHE compared to isogenic WT. Comet assay in V79 CHE-exposed cells showed no DNA damage. No mutations induced in CHE-TA98 and TA100 strains of S. typhimurium (with or without metabolic activation) was observed in the yeast strain XV185-14c CHE-exposed failed to induce mutations. CHE non-recombinogenic events induced in the diploid yeast strain XS2316. Absence of genotoxicity and demonstration of a non-toxic potential suggest that CHE, or some substances contained in it, may hold promise for a biotechnological application.
Keywords: cocoa, comet assay, genotoxicity, transition metals
Cacao (Theobroma cacao L.), a tropical evergreen tree belonging to the Malvaceae family.1 is the most economically important species of the genus Theobroma for many small crop farmers in developing countries with tropical climate. Cacao seeds (beans) constitute the raw material for the production of chocolate and confectioneries worldwide and some of their components are also used in cosmetics and pharmaceuticals.2
Cacao is produced in monocultures with up to 1,000 tress per hectar and this creates two problems: (a) phytopathogens like the basideomycete fungus Moniliophthora perniciosa, the causal agent of witches’ broommay cause dramatic losses in crop production and (b) the processing of the ripe cacao pods on the farm leaves a huge amount of organic waste in form of the empty pod husks.3-5 Some million tons of cacao pod husks are disposed of every year, often left behind in the fields, resulting in an undesirable waste of organic material.6 that contains valuable organic compounds. The simplest way to get rid of this pod husks is to macerate them and distribute them under the trees as organic fertilizer. This, however, may also spread plant pathogens, especially microbes that thrive on the rich organic material.7
Alternative approaches have been to extract cacao metabolites of economic value.8 from the pod husks.5,9,10 or to subject the macerated pod husks to aerobic fermentation that curbs the development of pathogenic microbes and produces a stable compost (biofertilizer) with antifungal activity that can be safely spread on the cacao farm.11
This aerobic fermentation of pod husks also produced a liquid water extract with antifungal activity. We standardized the extraction procedure and verified the extract’s antimicrobial potential.4 Here we wish to report our experiments that evaluated the biological safety. i.e., cytotoxic and genotoxic activities, of cacao husk extract (CHE).
Materials, chemicals, reagents and media
All plastics and media were purchased from Gibco®, Eppendorf® (Hamburg, Germany) and Becton, Dickinson and Company® (BD, Franklin Lakes, N. J., USA). All chemicals and reagents were P.A. grade and purchased from Sigma Aldrich Inc, Chemicals and Reagents (Sigma-Aldrich Co. LLC, USA) or from Merck Chemicals & Reagents (Merck KGaA, Darmstadt, Germany).
Preparation of Cacao pod husk extract - CHE
T cacaoplants selected for this study were Scavina-6 clones resistant against the fungus Moniliophthora perniciosa (formerly Crinipellis perniciosa (Stahel) Singer) growing in experimental areas of the Comissão Executiva de Planejamento da Lavoura Cacaueira (Executive Committee of the Plan of Cocoa Farming, CEPLAC) in Ilhéus, Bahia. Extract of T. cacao was prepared at Santos et al.4 as described.
Two g of this crude extract was reduced to powder using a porcelain mortar, suspended/dissolved in 10 mL of distilled water and filtered through a paper filter (Whatman, 80g/m2, porosity 3µm). The remaining non-dissolved material was re-submitted to the same procedure twice to yield a total of 30mL water-dissolved extract. The still remaining insoluble residue was resuspended 5mL of distilled water and subjected to sonication (Ultrasonic processor Gex 130, 130 W), with 10 pulses of 40 s each, 60% output with 10 s intervals, and filtered as described above. A total of 35mL of this aqueous extract was frozen at -20 °C, later concentrated by freeze-drying for 4 d in order to obtain cocoa husk extract .CHE (yield of 72.95% w/w). 100 g of CHE was submitted to biological analysis.
Analytical methods
Zinc (Zn), copper (Cu), manganese (Mn), iron (Fe) concentrations were determined by nitropercloric digestion according to the Association of Official Analytical Chemists (AOAC) guidelines.12 All analyses were carried out in triplicate, the results were expressed as mg/1000g.
Assays with S. cerevisiae
Strains and media: Relevant genotypes of yeast strains used in this work are given in Table 1. Media and solutions were prepared according to Burke et al.13 Complete liquid medium (YPD) was used for routine growth of yeast cells. For plates, the medium was solidified with 2% agar. Minimal medium (MM) was supplemented with the appropriate amino acids and nitrogen base uracil to yield synthetic complete medium (SC). For mutagenesis in the strain XV-185-14c, the omission media lacking lysine (SC-lys), histidine (SC-his), or homoserine (Sc-hom) were used. For evaluation of recombinogenisis, leucine was omitted from the synthetic complete medium (SC-leu) or supplemented with 0.2 % (w/v) cycloheximede (SC + cyh).
|
Strain |
Genotype |
Protein lacking |
Source |
|
Yeast strains |
|||
|
DY1457 |
Mata: ade6 can1Δ100oc his3 leu2 trp1 ura3 |
none |
David J Eide |
|
DEY1394 |
Like DY1457 except fet3Δ::His3 |
Class of integral membrane multicopper oxidases |
David J Eide |
|
DDY33 |
Like DY1457 except fet4Δ::Leu2 |
Low-affinity Fe(II) transporter of the plasma membrane |
David J Eide |
|
DEY1524 |
Like DY1457 except aft1Δ::Trp1 |
Transcription factor involved in iron utilization and homeostasis |
David J Eide |
|
ZHY6 |
Like DY1457 except zap1Δ::Trp1 |
Zinc-regulated transcription factor |
David J Eide |
|
BY4741 |
Matα his3Δ1 leu2Δ0 lys2Δ0 ura 3Δ0 |
none |
EUROSCARF |
|
Ntg2 |
Like SJR753 except ntg2Δ::hisG |
DNA N-glycosylase and apurinic/apyrimidinic (AP) lyase; involved in base excision repair |
RL Swanson et al. 1999 |
|
ΔFre1 |
Like BY4741 except fre1Δ::His3 |
Ferric reductase and cupric reductase |
From Dennis J Thiele |
|
ctt1Δ |
Like SOD+ exceptctt1Δ::TRP1 |
Cytosolic catalase T |
E.B.Gralla, Los Angeles |
|
|
|
|
|
|
XV185-14c |
MATα: ade2Δ2 his1-798 lys1Δ1 trp5-48 hom3-10 arg4-17 |
none |
Von Borstel et al.26 |
|
XS2316 |
MATα + leu1-1 trp5-48 +++ his1-208 |
none |
Machida & Nakai 1980 |
|
MATα ade6 leu1-12 + cyh2 met13 lys5-1 his1-208 |
|||
Table 1 Yeast strains used and their relevant genotypes
Yeast growth and treatments: Stationary phase (STAT) cultures were obtained by inoculation of an isolated colony in liquid SC for 48h. Exponential growth (LOG) were obtained by inoculation of 5x105 cells/mL of a SC culture in stationary phase into fresh 50 mL SC medium.
For all the experiments the cells in STAT phase or LOG phase, were washed twice with saline solution (0.9% NaCl, pH 5.0) and suspension of 2 x108 cells/mL was incubated at 30 °C for a time 24h with various concentrations of CHE in saline. To evaluate sensitivity to CHE, yeast cultures were exposed to concentrations verging from 5 to 40mg/mL. To verify mutagenicity and recombinogenisis, cells were pretreated with non-cytotoxic concentrations of CHE. After CHE exposure cells were washed in saline, plated in appropriate dilution for phenotypic analysis of determination survival or mutagenesis or recombinogenisis.
Sensitivity assay in yeast wild and mutant isogenic strains: Post-treatment, the cells were diluted in 1:10 steps in sterile saline and 5µL of each dilution (suspensions containing from 107 to 103cell/mL) was placed on surface of solid SC solid medium. Cellular growth on SC was determined after 5d incubation at 30°C. Photos were taken using a Canon Power Shot G10 camera. Photos represent one of at least three independent experiments with similar results.
For survival determination, suitable aliquots were plated in triplicate on solid SC, plates were incubated at 30 °C for 2-3 days before counting the colonies. Presented results are the mean of at least three independent experiments and error bars represent standard deviation as calculated by GraphPad Prism® program.
Detection of induced mutation and mitotic recombination: Haploid strain XV185-14c was used for assaying mutagenicity. After CHE exposure, cell survival was determined on SC (3-5 d, 30°C), and mutation induction (LYS, HIS or HOM revertants) on the appropriate omission media (7-10 d, 30 °C). Strain XS2316 (2 x 107 cells/mL) was used for analysis of mitotic recombination. After CHE treatment, appropriate dilutions of cells were plated onto SC plates to determine cell survival, and colonies grown on SC-leu and SC+cyh were scored for intragenic mitotic recombination (gene conversion) and intergenic recombination (crossing-over), respectively. Assays were repeated at least four times, and plating was performed in triplicate for each dose. CHE exposure concentration in mutagenesis assay was 0.5-2.mg/mL and 2 - 8mg/mL for recombinogenisis. Assays were repeated at least three times, and plating was performed in triplicate for each dose.
Salmonella/microsome mutagenicity assay
Salmonella typhimurium strains TA98 and TA100 were kindly provided by B. M. Ames (University of California, Berkeley, CA, USA). Mutagenicity was assayed by the preincubation procedure. The S9 metabolic activation mixture (S9 mix) was prepared according to Maron & Ames.14,15 Briefly, 100μL of test bacterial cultures (1-2x109 cells/mL) were incubated at 37 °C with different amounts of CHE in the presence or absence of S9 mix for 20 min, without shaking. Subsequently, 2mL of soft agar (0.6% agar, 0.5% NaCl, 50µM histidine, 50µM biotin, pH 7.4, 46 °C) were added to the test tube and poured immediately onto a plate of minimal agar (1.5% agar, Vogel-Bonner E medium, containing 2% glucose). Aflatoxin B1 (1µg/plate) was used as positive control for all strains in the presence of metabolic activation (with S9 mix). In the absence of metabolic activation, 4-nitroquinoline-oxide (4-NQO, 0.5µg/plate) was used for strain TA98 and sodium azide (1µg/plate) for strain TA100. Plates were incubated in the dark at 37 °C for 48 h before counting the revertant colonies.
CHE cytotoxicity
V79 cells were maintained in tissue-culture flasks at 37 °C in a humidified atmosphere containing 5% CO2, and were harvested by treatment with 0.15% trypsin and 0.08% of EDTA PBS. Cells (2 × 105 cells) were seeded into each flask and grown/incubated 1 d prior to treatment.
The concentrations used for each sample were based on trypan blue exclusion, which was determined as described by Robichova & Slamenova.16 Treated and washed, as well as control V79 cells were trypsinized, stained with trypan blue (0.4%), and the number of viable (uncolored) and dead (colored) cells was counted. The ratio of number of viable cells/all cells results in the percentage of viable cells. Dose was considered cytotoxic when cell survival was <70%.
Comet assay
The Comet Assay was performed based on the protocol proposed by Singh et al.17 Briefly, a cell suspension of 20μL of cell culture in 1mL of RPMI 1640 was exposed to 20μL of CHE (final concentration of 0, 625 to 5μg/mL, 1 h, 37 °C), 15μL of that cell suspension were mixed with 90μL of low-melting agarose, in a micro-centrifuge tube. The cell suspension was transferred to an agarose pre-coated glass slide, covered with a glass cover slip (22mm x 66mm), and immediately placed in a refrigerator (4 °C) for 5 min to allow complete agarose solidification. Slides were prepared in duplicate and coded.
Cover slips were then removed and the slides immersed in cold (4 °C) lysing solution (2.5 M NaCl, 100 mM Na2EDTA, 10 mM Tris with 1% (v/v) Triton X-100 and freshly added 10% DMSO pH 10), protected from light for at least 8 h. Cells were exposed to alkali (300 mM NaOH/1 mM Na2EDTA, pH >13, 30 min, 4 °C), to allow DNA unwinding and expression of alkali-labile sites. For electrophoresis, an electric yield of 78 V/cm was applied for 25 min at 4 °C. After electrophoresis, the slides were neutralised and silver stained.18 One hundred cells were scored visually according to the tail length and the amount of DNA present in the tail. Each comet was given an arbitrary value of 0-4 (0, undamaged; 4, maximally damaged), as described by Collins et al.19 Damage score was thus assigned to each sample and can range from 0 (completely undamaged: 100 cells x 0) to 400 (with maximum damage: 100 cells x 4). International guidelines and recommendations for the comet assay consider that visual scoring of comets is a well-validated evaluation method as it is highly correlated with computer-based image analysis.19,20
Statistical analysis
The results were analyzed by the Salmonella Statistic Assay (Environmental Monitoring System Laboratory, EPA - Software Version 2.3, April 1988). A test substance was considered mutagenic when significant dose-response and ANOVA variance were observed, and the increase in the mean number of revertants on test plates was at least twice higher than that observed in the negative control plates.
In order to know the interaction of CHE with S. cerevisiae, we performed sensitivity experiments. The range of lethal CHE concentrations on yeast was determined in a drop test using different haploid strains. The mutant strain fet3Δ showed high sensitivity to 10mg/mL CHE exposure as compared to the WT control, whereas mutant alleles fre1Δ and ntg2Δ) did not confer sensitivity to the same CHE exposure (Figure 1).
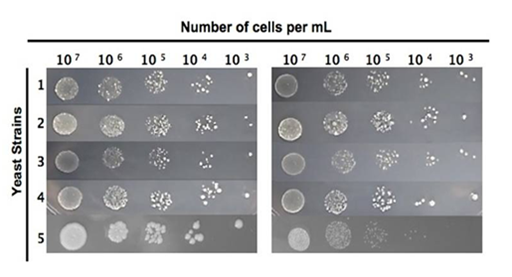
Figure 1 Sensitivity of haploid yeast strains to CHE.
Yeast strains: A: control, B: CHE exposure (condition 24 h, 30 oC). 1. BY4741; 2. ctt1∆; 3. fre1∆;4. ntg2∆; 5. fet3∆.
In order to quantify drop test results for CHE-sensitive strains, we determined survival kinetics of haploid yeast strains. We observed that STAT yeast cells harboring fet3Δ and aft1Δ mutant alleles were sensitive to 10 mg/mL CHE as compared to the isogenic WT that showed some 85% survival, while fet4Δ and zap1Δ mutants had WT-like survival (Figure 2). Several transition metals, such as zinc (Zn), iron (Fe), manganese (Mn), or copper (Cu) are essential micronutrients for all organisms, used in redox processes, regulation of osmotic pressure, and also are essential components of enzymes .21. However, transition metals in high concentration can damage the structure of DNA, cell membranes and may change enzymes specificity, causing oxidative deterioration of biological macromolecules.22,23 We found that CHE contains a high amount of Fe, followed by Mn and Zn, with intermediate proportions of Cu (Table 2). In theory, the most simple and effective cellular strategy to avoid toxic intracellular concentrations of metals is to eliminate the channels or transporters responsible for their uptake by the membrane.24 Metals may enter into the cytoplasm through multiple transporters; however the absence of even only one of these transport routes may already disturb the normal metal metabolism of the cells.25
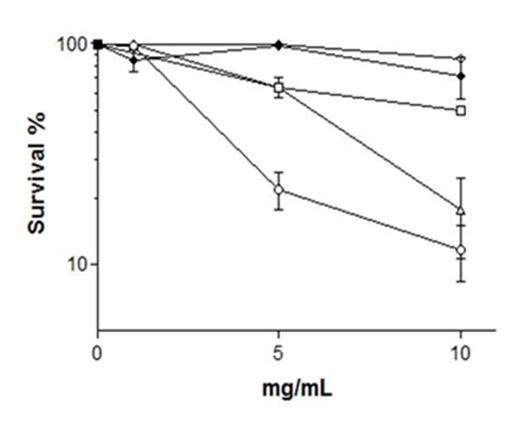
Figure 2 Sensitivity to CHE of haploid STAT yeast cells.
Cells were diluted in saline. (L) WT; (♢)zap1Δ (◯)fet3Δ; (☐)fet4Δ (∆)aft1Δ
|
Minerals |
(mg/dm-3) |
|
Fe |
1155±19.1 |
|
Zn |
134±2.5 |
|
Cu |
58.6±1.1 |
|
Mn |
292.1±1.3 |
Table 2 Chemical composition of extract of T. cacao
Results are the mean of triplicate measurements ± SD.
Since it was unknown if CHE interacted with DNA we chose the unicellular eukaryotic organism S. cerevisiae to determine whether CHE produce DNA damage and whether this induces mutagenesis and recombination.
The haploid yeast strain XV185-14c was used for assaying mutagenicity, as it permits the detection of two types of locus-specific (reversion of the lys1-1 ochre or his1-7 missense alleles) and frame shift mutations (hom3-10) .26. Our experiments clearly show that CHE was not able to induce mutations (in concentrations of 50 to 200μg/mL), neither in alleles allowing locus-specific mutation nor to frame shift sensitive allele hom3-10; did it also not significantly reduce cell viability as compared to the negative control (Table 3). The percentage of survival among the different treatments ranged between 78.9 and 99.8% compared to the negative control. Only the positive control treatment with 4NQO 0.5μg/mL was significantly different, with 65.5% survival.
|
Agent |
Treatment (µg/mL) |
Survival (%) |
His1/107 Survivorsa |
Lys1/107 Survivorsb |
Hom3/107 Survivorsb |
|
Stationary phase - Cells treated in NaCl 0,9% |
|||||
|
4NQOd |
0.5 |
65.1±2.4 |
48.2±10.9*** |
11.6±3.4 |
4.2±0.9 |
|
|
|||||
|
NCc |
0 |
100e |
9.9±4.0e |
4.2±0.8e |
3.0±0.9e |
|
WSC |
50 |
92.3±3.7 |
10.8±3.4 |
3.9±0.9 |
2.1±0.5 |
|
|
100 |
80.0±7.7 |
11.4±4.0 |
4.1±0.7 |
2.3±1.2 |
|
|
200 |
78.9±8.3 |
9.8±3.0 |
2.8±0.7 |
2.1±0.4 |
|
Exponential phase - Cells treated in NaCl 0,9% |
|||||
|
4NQO |
0.5 |
99.1±1.72 |
52.2±3.8*** |
16.6±0.1 |
6.2±0.7 |
|
|
|||||
|
NC |
0 |
100 |
14.0±0.2 |
2.6±0. 4 |
0.9±0.25 |
|
WSC |
50 |
97.6±3.3 |
18.0±0.6 |
2.5±0.07 |
2.2±0.07 |
|
|
100 |
99.8±0.2 |
13.0±0.1* |
2.2±0.02 |
3.3±0.5 |
|
|
200 |
99.1±0.2 |
7.0±0.4 |
0.8±0.02 |
1.9±0.9 |
Table 3 Indution of point mutation (his1-7), Ochre allele (lys1-1) and frameshift (hom3-10) mutations in haploid XV185-14 strain of S.cerevisiae after CHE treatments aLocus-specific revertants; bLocus-nonspecific revertant (forward mutation); cNegative control (solvent); dPositive control; eMean and standard deviation per three independent experiments *Significantly different from the negative control at P<0.05; **P<0.01; ***P<0.001/ one-way ANOVA-Tukey’s multiple comparison test
Possible recombinogenic effects of CHE were investigated by using the diploid yeast strain XS2316, which allows the detection of two-forms of mitotic recombination (crossing-over and gene conversion). The recombinogenic effect of CHE was assayed using diploid LOG cells held under either growth or non-growth (saline) conditions (Table 4). CHE-exposed cells were grown on SC medium, yielding results of cell survival. Colonies grown on SC-leu and SC+cyh were scored for intragenic recombination (gene conversion) and intergenic recombination (crossing-over), respectively. CHE did not induce any recombinogenic events in diploid LOG cells (Table 4). However, we observed an increase in mitotic crossing-over for almost all doses of CHE-exposure in STAT cells in relation to negative control. It is possible that this increase does not reflect a consequence for cells in mitotic crossing-over, per se, but may be a secondary effect of recombination between homologous DNA duplexes. In mitotic cells, homologous recombination provides a means for repair chromosomes to their original state after breaks during replication, or from exogenous insults.27 In order to eliminate possible revert ants to cycloheximide-resistance resulting from mutation at the CYH2 locus, resistant colonies were replica-plated on a series of plates with SC-Lys, SC-Met and SC-Ade media for screening the recombination of cyh2 homozygocity.
|
Agent |
Treatment (µg/mL) |
Survival (%) |
Crossing-over/105 Survivorsa |
Gene Conversion/105Survivorsb |
|
Stationary phase - Cells treated in NaCl 0,9% |
||||
|
4NQOc |
0.5 |
97.5±3.5 |
106.8±2.5 |
44.3±2.9 |
|
|
0 |
100e |
17.7±0.3e |
2.8±1.5e |
|
WSC |
200 |
99.5±0.6 |
35.4±4.2 |
1.7±1.5 |
|
|
400 |
95.6±6.2 |
36.4±4.3 |
3.6±1.4 |
|
|
800 |
95.0±7.0 |
40.9±3.8 |
1.1±0.7 |
|
Exponential phase - Cells treated in NaCl 0,9% |
||||
|
4NQO |
0.5 |
84.3±1.7 |
153.8±2.5 |
39.1±6.1 |
|
NC |
0 |
100 |
12.3±0.9 |
4.2±2.4 |
|
WSC |
200 |
99.5±0.7 |
18.0±0.5 |
5.2±3.6 |
|
|
400 |
79.3±23.3 |
26.0±0.2 |
6.1±1.7 |
|
|
800 |
89.7±3.1 |
22.2±2.2 |
2.9±0.5 |
Table 4 Effects of CHE in diploid XS2316. Induction of crossing-over (+/cyh2) and gene conversion (leu-1/leu1-12)
aMean and standard deviation per three independent experiments; bNegative control (solvent); cPositive control
Possible mutagenicity in prokaryotes exposed to CHE was determined in the Ames test using S. typhimurium strain TA98 that detects frame shift mutations based on the reversion of S. typhimurium strain from His− to His+ (Table 5). The CHE dose range was determined in a range finder experiment with strain TA100 with and without metabolization, and cytotoxicity was not observed at concentrations up to 5,000μg/plate (data not shown). In mutagenicity assays CHE doses ranging between 1,000-5,000 μg/plate were used. The extract was not mutagenic in strain TA98 (detects frame shift mutation in the DNA target -C-G-C-G-C-G-C-G) in the absence or presence of metabolic activation. Also, no mutagenicity was seen in strain TA100 detecting base pair substitutions (Leucine .GAG by Proline .GGG) in the absence or presence of metabolic activation. Results therefore indicated that CHE in exposure doses up to 5 mg/mL was not mutagenic in the Salmonella/microsome assay.
|
S. typhimurium strains |
|||||
|
Agent |
Concentration |
TA100 |
|
TA98 |
|
|
|
|
Rev/platea |
MIb |
Rev/platea |
MIb |
|
Without metabolic activation (-S9) |
|
|
|
||
|
NCc |
- |
125.3±4.6 |
- |
22.3±4.5 |
- |
|
Extract |
1000 |
104.3±6.4 |
0.83 |
26.0±2.0 |
1.17 |
|
|
2000 |
101.0±14.8 |
0.81 |
20.7±3.5 |
0.93 |
|
|
3000 |
103.7±11.7 |
0.83 |
22.0±4.4 |
0.99 |
|
|
4000 |
112.3±3.5 |
0.90 |
19.3±1.5 |
0.87 |
|
|
5000 |
114.3±13.0 |
0.91 |
18.3±9.0 |
0.82 |
|
PCd |
0.5 (4NQO) |
468.0±46.9*** |
3.74 |
182.3±30.6*** |
8.18 |
|
With metabolic activation (+S9) |
|
|
|
||
|
NCc |
- |
156.3±13.2 |
- |
23.7±1.5 |
- |
|
Extract |
1000 |
157.7±12.7 |
1.01 |
22.3±0.6 |
0.94 |
|
|
2000 |
161.0±15.4 |
1.03 |
27.0±4.4 |
1.14 |
|
|
3000 |
160.3±5.3 |
1.03 |
22.5±6.4 |
0.95 |
|
|
4000 |
157.0±23.1 |
1.01 |
21.7±5.9 |
0.92 |
|
|
5000 |
152.0±8.0 |
0.97 |
27.0±6.6 |
1.14 |
|
PCd |
1 (AFB1) |
454.0±144.8* |
2.90 |
764.7±130.1*** |
32.3 |
Table 5 Induction of his+ revertants in S. typhimuriumstrains by CHE with and without metabolic activation (S9 mix)
aNumber of revertants/plate: mean of three independent experiments ± SD; bMI: mutagenic index (nº. of his+ induced in the sample/nº. of spontaneous his+ in the negative control); cNC: negative control distillated water (10 µl) used as a solvent for the extract; dPC: positive control (-S9) sodium azide to TA100; 4-NQO to TA98; (+S9) aflatoxin B1.
*Data significant in relation to negative controlp<0.05; *** p<0.001
The ability of CHE to damage DNA in mammalian cells was estimated by the Single Gel Electrophoresis Test (Comet assay) in which the tail length (the comet) indicates the extent of migration of the genetic material in the direction of the anode is giving an estimate of DNA damage, particularly of the extent of DNA strand breaks is CHE exposure concentrations of up to 10ug/mL did not reveal significant DNA breakage (genotoxicity) (Figures 3A & 3B) as comets were not significantly altered as compared to the positive control. Our results not demonstrate the genotoxic and comutagenic effects of CHE in V79 cells. DNA lesions leading to effects in the comet assay can be strand breaks which may be relevant to the DNA modifications such as basic sites with relevance to the induction of gene mutation.28, however, small lesions may be repaired Without resulting in genetic changes. Thus, the conclusions regarding the mutagenicity of a assay substance are not made solely on the basis of the effects of the comet assays.
In summary, our results show that CHE in the applied exposure concentrations was not genotoxic in our pro- and eukaryotic microbial and mammalian test systems.
Research financed by CNPq. R.X.S. and D.O. held CNPq fellowships. G.G. is grateful to CNPq and CAPES for financial support. M.B. is visiting professor at UESC.
The authors declare that there is no conflict of interests.
None.

©2017 Santos, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.