International Journal of
eISSN: 2381-1803


Research Article Volume 12 Issue 6
NIS Labs, USA
Correspondence: Gitte S Jensen, Research Director, NIS Labs, 1437 Esplanade, Klamath Falls, Oregon 97601, USA, Tel (541) 884-0112
Received: September 21, 2019 | Published: November 19, 2019
Citation: Benson KF, Jensen GS. Bacteria in blood from fibromyalgia patients include the Aquabacterium genus, producing metabolites with inflammatory properties in vitro. Results from a pilot study. Int J Complement Alt Med. 2019;12(6):232?239. DOI: 10.15406/ijcam.2019.12.00479
Purpose: To evaluate bacterial presence and evidence of inflammatory metabolic activity in blood of patients with Fibromyalgia Syndrome (FMS).
Patients and methods: A human clinical pilot study involved eight FMS patients and four healthy controls. Whole blood and purified erythrocytes were cultured and examined by darkfield fluorescence microscopy and flow cytometry. DNA was subjected to amplification using polymerase chain reactions. Bioactivity of bacterial metabolites was tested on peripheral blood mononuclear cells (PBMC), cultured in the presence of metabolite fractions from FMS and healthy control erythrocyte cultures. CD69 and CD25 expression was evaluated by flow cytometry, and levels of inflammatory cytokines tested by Luminex array.
Results: Blood cultures from fibromyalgia patients revealed growth of bacteria under culture conditions involving RPMI-1640 medium, increased levels of glucose and iron, and reduced oxygen. The ex vivo expansion of bacteria through culture, combined with 16S rDNA PCR analysis of DNA isolated from whole blood and erythrocyte cultures, identified bacterial sequences derived from the Aquabacterium genus. The design of Aquabacterium-specific primers resulted in detection of Aquabacterium DNA in blood from some healthy control subjects as well, suggesting a low level of bacteria in a more quiescent state. PBMC cultured in the presence of supernatants from FMS erythrocyte cultures showed increased levels of CD69+ CD25+ lymphocytes, and increased production of the inflammatory cytokines IL-6 (p<0.25), IL-8 (p<0.08), and TNF-alpha (p<0.01) when compared to supernatants from erythrocyte cultures from healthy individuals.
Conclusion: Bacteria from the biofilm-forming microaerophilic Gram-negative Aquabacterium genus were detected in blood from FMS patients and some healthy controls. Only bacterial cultures from FMS patients produced inflammatory metabolites, suggesting that Aquabacterium bacteria from FMS patients may express different genes than in healthy controls, potentially contributing to the chronic pain and inflammation characteristic of FMS.
Keywords: biofilm, cell wall-deficient, chronic inflammation, immune activation, l-form bacteria, cytokines
DNA, deoxyribonucleic acid; FMS, fibromyalgia syndrome; IL-1, interleukin-6; IL-8, interleukin-8; PBMC, peripheral blood mononuclear cells; PCR, polymerase chain reaction; TNF-α, Tumor necrosis factor-alpha; WB, whole blood
Fibromyalgia syndrome (FMS) is a chronic musculoskeletal pain disorder with unknown cause.1 Chronic diffuse pain, stiffness, multiple tender points, fatigue, poor sleep, and increased pain response to pressure (allodynia) are classic symptoms of FMS.2 FMS affects approximately 2% of the population and is more common in women with a ratio of about 7:1 between women and men. Current research points to both central and peripheral nervous system dysfunctions including altered nociceptor sensitivity, as well as systemic inflammation,3 including increased levels of pro-inflammatory cytokines such as Tumor Necrosis Factor-α (TNF-α ).4 FMS has symptomatic overlap with another chronic pain disorder, Chronic fatigue syndrome (CFS). It has been postulated that both FMS and CFS could have underlying microbial etiologies.5,6 Furthermore, similarities in symptoms between FMS and CFS and “sickness behavior” implicate an immune component.7-9 Sickness behavior results from the release of proinflammatory cytokines by leukocytes in response to bacterial lipopolysaccharide and other toxins and leads to low-grade systemic inflammation which has been implicated in chronic pain, depression, and fatigue,10,11 the hallmark symptoms of FMS and CFS.
Bacteria are omnipresent in the environment, including the human body, where they outnumber our own cells by an order of magnitude. These communities of commensal bacteria colonizing the human body are referred to as “microbiome”.12,13 Humans are dependent on beneficial bacteria for proper development of the immune system and many aspects of health, including protection from pathogens. However, for this type of symbiosis to be successful, it is important that a balance be maintained between these commensal bacteria and human physiology. Benign bacteria can become pathogenic when the proper ratio is no longer maintained or in situations of immune suppression that exist in the conditions of cancer, AIDS, cystic fibrosis, and chronic granulomatous disease. In these situations, pathologies such as dysbiosis, metabolic syndrome, inflammatory bowel disease, skin pathologies, periodontal disease, and other disorders can manifest.14 These commonly existing, but typically benign, bacteria also add to the complexity of detecting and identifying bacteria of a more pathological nature, as these may coexist in biofilms serving as reservoirs of pathogenic organisms, and potentially facilitating gene transfer that may enhance the microbial resistance to oxidative stress, and reduce the recognition by the immune system of the host.15,16
Commensal bacteria are well-known as essential residents of the skin, nasal cavity, vagina, and gastrointestinal tract – anywhere the body has contact with the outside environment. Very recent research has provided evidence for the widespread existence of bacteria in healthy blood of both animals17,18 and humans. This blood microbiome shows differences in bacterial species make-up in different blood fractions19 and erythrocytes contain unique bacteria as well as a higher proportion of pathogenic bacteria, many of these belonging to the Proteobacteria phylum. Proteobacteria make up a much larger percentage of the blood microbiome (>80%) compared to the gut microbiome where Firmicutes and Bacteroidetes are the predominant phyla. Furthermore, Proteobacteria have been implicated in dysbiosis and diseases of the gastrointestinal tract,20-22 and are increased in the gut microbiome of chronic fatigue syndrome,23 while perturbations of the blood microbiome have been linked to disease24 including diabetes,25 cardiovascular disease,26 and liver fibrosis.27
The presence of occult and often unculturable bacteria in human blood is challenging to document, but recent elegant advances in molecular biology have led to definitive characterization of the human blood microbiome.28 Bacterial 16S rRNA gene analysis is the method of choice for its sensitivity and unequivocal interpretation. This method may be used as a culture-independent tool or applied to cultured samples where ex vivo growth may have amplified the number of bacteria for easier detection of low numbers of bacteria. Originally used for the query of environmental samples, this method has driven the explosion in the recent exploration of the microenvironment of the human body. In addition to a gut microbiota, microbiota of the skin, lung, oral mucosa, vagina, seminal fluid, conjunctiva, and blood have been identified.29–35
Recent advances have allowed optimization of 16S rDNA PCR methods designed for the analysis of clinical samples, including blood.36–38 In parallel, an ever-increasing number of known bacterial genomes are available in databases. These factors in combination may help demystify a large volume of previous microbiology findings, in which a discrepancy between microscopy observations and detection of bacterial DNA has become increasingly obvious.39 However, even with the objectivity of modern molecular genetics methods, the detection of low numbers of bacteria in human blood or tissue presents many technical challenges.40–42 The ability of such bacteria to establish cell-wall deficient intracellular presence, possibly in the form as biofilms, adds to the challenge of documenting presence and bioactivity of intraerythrocytic microbes.43,44
Previously, we monitored erythrocytes of a patient diagnosed with FMS during several years of escalating chronic illness.45 Bacteria could consistently be cultured from the erythrocyte fraction, and the successful culture correlated with disease activity, suggesting a higher level of pathogenic activity at those times, and we hypothesized that secreted metabolites interfered with the host immune system. Bacterial DNA was isolated by 16S rDNA PCR, a 465bp PCR product sequenced and the resulting sequence showed 100% homology with a group of soil bacteria known as alpha-proteobacteria (GenBank accession AF399924.1).
Based on our initial finding, we designed the pilot study reported here. The results from the pilot study presented here is a first step to provide more systematic documentation of differences in blood microbiota between FMS patients and healthy controls. An important part of this work was to explore whether the intraerythrocytic bacteria, actively growing in vitro, could secrete metabolites that interacted with host immune cells in an inflammatory manner.
Reagents
The following reagents were obtained from Sigma-Aldrich (St. Louis, MO): Histopaque 1077 and 1119, phosphate-buffered saline (PBS), lipopolysaccharide (LPS), interleukin-2 (IL-2), Roswell Park Memorial Institute 1640 medium (RPMI-1640), penicillin-streptomycin 100X, fetal calf serum (FCS), 1000X iron supplement, and Luria broth. The 6X gel loading dye and SYTO® 9 DNA dye were from Thermo Fisher Scientific (Waltham, MA).
Study design
The methodology work reported here involved a clinical pilot study involving 12 people (8 FMS, 4 controls) with evaluation of bacterial DNA in whole blood, freshly isolated erythrocytes, and erythrocytes after seven days of culture ex vivo. Supernatants from the erythrocyte cultures were used for the testing of potential effects of secreted bacterial metabolites on the human host (described below). For the clinical pilot study, blood samples from eight FMS patients and four healthy controls were obtained following written informed consent, as approved by the Sky Lakes Medical Center Institutional Review Board, (FWA 2603). FMS patients were all female, ranging in age from 25 to 62years (mean 48years) and duration of FMS ranging from 1 to 25years (mean 15 years). Age, gender, and disease duration for these participants are summarized in Table 1.
Vol # |
Gender |
Agea |
Durationb |
16S universal primers |
16S Aquabacterium primers |
FMS01 |
F |
42 |
21 |
WB-D7c, E-D7d |
F-WBe, E-D7 |
FMS02 |
F |
55 |
26 |
WB-D7, E-D7 |
WB-D7 |
FMS03 |
F |
25 |
3 |
||
FMS04 |
F |
32 |
1 |
WB-D7 |
WB-D7 |
FMS05 |
F |
56 |
24 |
WB-D7 |
|
FMS06 |
F |
58 |
15 |
WB-D7 |
|
FMS07 |
F |
56 |
13 |
WB-D7 |
WB-D7 |
FMS08 |
F |
62 |
16 |
WB-D7 |
|
CTRL01 |
F |
59 |
WB-D7, E-D7 |
WB-D7 |
|
CTRL02 |
F |
48 |
WB-D7 |
||
CTRL03 |
M |
49 |
|||
CTRL04 |
M |
52 |
WB-D7 |
Table 1 Comparison of 16S rDNA PCR results using universal and Aquabacterium-specific primers
aAge of participants in years; bDuration of FMS in years; cWB-D7, Whole blood 7-day cultures with positive PCR results; d E-D7; Erythrocyte 7-day cultures with positive PCR results; e F-WB, Freshly drawn whole blood
Erythrocyte isolation and culture
Peripheral venous blood was obtained as follows: The skin surface of the antecubital region was swabbed continuously for 30 seconds with betadine and allowed to dry. Peripheral venous blood was drawn by venipuncture of the antecubital vein and collected into a single vacutainer tube containing heparin (Becton Dickinson, Franklin Lakes, NJ), and layered onto a Histopaque 1077/1119 double gradient and centrifuged at 2400rpm for 25minutes (Centra-MP4, IEC, Needham Heights, MA). Erythrocytes were isolated and washed four times with phosphate buffered saline using sterile technique. Erythrocyte cultures were established by placing 200µl of packed erythrocytes into 10 mL of RPMI-1640 medium supplemented with iron (2X final concentration), and cultures maintained at 37oC, 5% CO2 for 7 days. The processing of the 12 blood samples occurred over a total of three days. On each day that blood samples were processed, both fibromyalgia and control samples were processed in parallel. Following 7 days’ incubation, erythrocyte cultures were centrifuged and the fraction containing cells, cell fragments, and bacteria was isolated by centrifugation, and the culture supernatant (metabolite fraction) harvested (Figure 1).
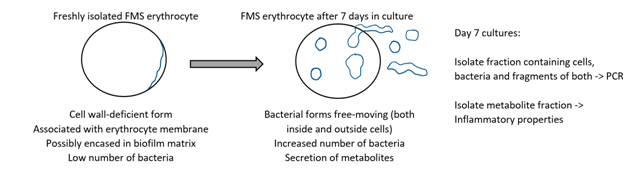
Figure 1 Diagram of the methodological approach to expanding bacteria by ex vivo culture, followed by PCR on the cell fraction, and harvesting of metabolites to examine impact on inflammation.
Microscopy
Whole blood, as well as fresh and cultured fractions thereof, were examined by dark-field microscopy. For fluorescence microscopy, a mixture of 10µl of SYTO® 9 (Thermo Fisher Scientific, Waltham, MA) and 10µl erythrocytes was incubated in the dark for 10 minutes. Combined darkfield and fluorescence microscopy was performed using a CytoViva® enhanced darkfield optical microscope with high resolution hyperspectral imaging (CytoViva Inc., Auburn AL, USA).
DNA extraction from human whole blood and bacterial control strains
DNA was prepared from whole blood, freshly isolated erythrocytes, cultured erythrocytes, and from erythrocyte culture supernatants. When processing DNA from cultured erythrocytes and from erythrocyte culture medium, 1µg of carrier RNA (Qiagen, Valencia, CA) was added to the samples prior to DNA extraction.
DNA sequencing of 16S rDNA PCR products
PCR was performed on DNA isolated from whole blood (WB), erythrocytes, cultured erythrocytes, and medium from 7-day cultures. Amplification products obtained from 16S rDNA PCR were separated on a 1% agarose gel and purified using either QIAquick PCR purification kit (single DNA band on agarose gel, Qiagen, Valencia, CA) or QIAquick gel extraction kit (more than one visible DNA band on agarose gel – DNA from each band isolated separately, Qiagen, Valencia, CA). Amplification of DNA from Escherichia coli (E. coli) was used as a reference, and PCR amplification products that were in the same size range as the PCR product from E. coli DNA were physically excised from the agarose gel and the DNA isolated. Twenty nanograms of DNA template was combined with 25pg of primer and samples were sent to a commercial sequencing facility (Genewiz, South Plainfield, NJ). Sequencing data obtained from 16S rDNA PCR products from human blood samples was used to query the public gene sequence databases GenBank (NCBI, Bethesda, MD) and the Ribosomal Database Project (Center for Microbial Ecology, Michigan State University, East Lansing, MI).
Aquabacterium-specific 16S rDNA PCR
Once a positive match was made between the bacterial 16S rRNA gene sequence recovered from human blood samples and that of an Aquabacterium, Aquabacterium-specific primers were designed by comparing the Aquabacterium 16S rRNA gene sequence to that of E. coli and choosing regions where the two bacteria differed in sequence. As a positive control, genomic DNA isolated from the bacterial strain Methylibium petroleiphilum PM1 was obtained from ATCC (Manassas, VA). The following PCR conditions were used: 1X PCR buffer, 1.5 mM MgCl2, 0.4 mM dNTPs, 1.25 U Taq polymerase and 0.4 µM of primer in a total reaction volume of 25 µl. Five microliters of extracted DNA were used per reaction and reactions heated at 95oC for 4min., followed by 10 cycles of 94oC, 45 sec., 68oC, 30 sec., 72oC, 50 sec. and then 25 cycles of 94oC, 45 sec., 64oC, 30 sec., 72oC, 50 sec. with a final extension step of 72oC for 5 min. Primer sequences were Burk For4: 5’-GGCCGCAAGGTGCTGACGAGTG-3’ and Burk Rev4: 5’-CTGAACAGCAAGCCGTCCAAC-3’.
Evaluation of inflammatory properties of metabolite fraction
To evaluate the potential biological activity of bacterial metabolites in culture supernatants from the erythrocyte cultures, these were applied to cultures of peripheral blood mononuclear cells (PBMC) from a healthy donor. Heparinized peripheral whole blood from a healthy control (Study participant CTRL02) was layered onto a Histopaque double gradient as described above. The PBMC fraction was harvested and washed two times with sterile PBS. PBMC were then suspended at 10µ/mL in RPMI-1640 medium containing penicillin, streptomycin, and 10% fetal calf serum. Supernatants from 7-day erythrocyte cultures (stored as frozen aliquots) were diluted 1:4 with PBS and passed through a sterile 0.2µM cellulose acetate filter (Corning Inc., Corning, NY). Cultures were established by combining 180µl PBMC with 20µL of PBS (untreated cultures), or 20µL of 7-day erythrocyte culture supernatants from 4 FMS patients and 3 healthy controls (FMS01, FMS02, FMS04, FMS07, CTRL01, CTRL02, CTRL04), selected based on positive results from PCR using the universal primer. Cultures were performed both in the absence of and the presence of Interleukin-2 (IL-2, 100IU/mL). The highly inflammatory lipopolysaccharide (LPS) from E. coli was used as a positive control for immune cell activation (10ng/mL). Cultures treated with IL-2 were used as a control for cultures that received IL-2 along with 7-day erythrocyte culture supernatants. Cultures were established in quadruplicate and incubated at 37°C, 5% CO2 for 24 hours. After 24 hours, PBMC were stained for 15 minutes with the monoclonal antibodies CD69 fluorescein isothiocyanate and CD25 brilliant violet 421 (Becton Dickinson, Franklin Lakes, NJ) at the recommended concentrations. Flow cytometry was performed with an Attune acoustic-focusing flow cytometer (Thermo Fisher Scientific, Waltham, MA). Data analysis utilized gating on forward/side scatter to evaluate CD69 expression on lymphocyte, monocyte/macrophage, and neutrophil subsets. The lymphocyte subpopulation was further analyzed for CD25 and CD69 fluorescence to determine the percentage of CD69+CD25+ lymphocytes.
Cytokine profiles from PBMC cultures
Supernatants were harvested from PBMC cultures described above following 24-hour culture and the concentrations of IL-6, IL-8 (CXCL8) and TNF-α quantitated using Bio-Plex protein arrays (Bio-Rad Laboratories, Inc., Hercules, CA) and utilizing xMAP® technology (Luminex, Austin, TX, USA).
Statistical analysis
Average and standard deviations for immune activation data were calculated using Microsoft Excel. Statistical analysis was performed using the 2-tailed, independent t-test. Statistical significance was p<0.05, and a high level of significance p<0.01.
Bacterial association with blood from FMS patients
Blood samples from FMS patients were examined by CytoViva combined darkfield and fluorescence microscopy, both on the day of the blood draw and after 7 days in culture. Whereas freshly isolated erythrocytes did not show evidence of bacteria, the cultured erythrocytes showed evidence of viable, motile organisms. The emerging bacterial forms showed association with the red blood cell membranes in the form of dense particles, and as free forms outside the erythrocytes (Figure 2B & 2D). There were also long rod-shaped forms outside the erythrocyte cells. There were some morphological differences when the erythrocytes were cultured in media with different nutrient profiles: Erythrocytes cultured in RPMI 1640 with added glucose and iron showed longer rod-shaped forms and also many extracellular dense particles (Figure 2B), whereas erythrocytes cultured in DMEM: F12 showed that some erythrocytes had tubular structures attached to the cell membrane but almost no extracellular bacterial forms (Figure 2D).

Figure 2 Erythrocyte cultures from fibromyalgia.
Emergence of microbial forms during ex vivo culturing of erythrocytes in different nutrient-rich media. CytoViva darkfield microscopy images of erythrocyte cultures from a fibromyalgia patient. Shown are typical micrographs from (A) Day 0, (B) Day 5 of erythrocytes cultured in phosphate buffered saline (PBS) supplemented with 17 mM galactose and (C) Day 0 and (D) Day 5 of RBC cultured in DMEM:F12 medium (without Hepes) 1:1 supplemented with 1X vitamins, 1X trace minerals, 2X iron, 0.000185% HCl and 100mM sucrose. After 5days of culture in PBS supplemented with galactose, RBC increased in size and cell membranes became very thin. In addition, RBC were filled with dense particles that were moving and tubular structures as well as moving particles were seen floating freely in the medium (B). After 5days of culture in supplemented DMEM: F12 medium, some erythrocytes had tubular structures attached to the cell membrane (D).
16S rDNA PCR on cultured blood samples
The results of 16S rDNA PCR only showed DNA amplification in fresh blood samples from 1 of the 12 donors, namely FMS patient FMS-01. In contrast, positive amplification of bacterial DNA was obtained from 7-day erythrocyte cultures from 5 of the 12 donors, comprising 4 FMS patients and 1 healthy donor who had a past medical history of symptoms associated with rheumatism (Table 1). A comparison of the sequence recovered from 16S rDNA PCR of FMS samples to 16S rRNA sequences deposited in the Ribosomal Database Project46 identified the sequence as derived from a bacterium belonging to the genus Aquabacterium, a member of the class beta-proteobacteria, order Burkholderiales that consists of both innocuous and pathogenic biofilm-forming bacteria, common in the environment.47
When the 7-day blood and erythrocyte cultures were tested using our 16S rDNA probe for Aquabacterium, this resulted in amplification of DNA from the same 4 FMS patients and the same healthy control. In addition, bacterial DNA was amplified from erythrocyte cultures of an additional 3 of the FMS patients, leaving only 1 of the 8 FMS patients (FMS-03) without detectable Aquabacterium DNA. Aquabacterium DNA was also amplified from blood cultures of two additional healthy controls. Table 1 compares the results that were obtained with 16S rDNA universal primer pair 27F/556R to the results obtained with the Aquabacterium-specific 16S rRNA PCR primers.
Increased numbers of activated lymphocytes and increased levels of inflammatory cytokines by FMS erythrocyte culture supernatants
To evaluate the possible production of inflammatory metabolic products from the intraerythrocytic bacteria, the erythrocyte culture supernatants were harvested as a source of such metabolites, sterile-filtered to remove any intact bacteria, and used for testing in a bioassay using peripheral blood mononuclear cells (PBMC) from a healthy donor, in the absence versus presence of Interleukin-2 (IL-2). The PBMC cultures were exposed to supernatants/metabolites from those 7-day erythrocyte cultures from 4 FMS patients that had showed detectable bacterial DNA using universal PCR primer as well as Aquabacterium PCR primer and 3 healthy controls that had showed detectable DNA using Aquabacterium PCR primer, (please refer to Table 1) for 24 hours, after which immune cell activation and cytokine production were measured. The numbers of the rare population of CD69+CD25+ activated lymphocytes increased in response to supernatants from all 7-day erythrocyte cultures (Figure 3A), however, in the presence of interleukin-2 (IL-2), this increase was much greater in cultures exposed to 7-day erythrocyte culture supernatants derived from FMS patients than those exposed to supernatants from healthy control subjects (Figure 3B). Due to the low number of subjects, statistical significance was not reached, but the results did approach a statistical trend (p<0.11). In the presence of IL-2, production of the proinflammatory cytokines IL-6, IL-8 and TNF-α were increased when exposed to supernatants from FMS patient 7-day erythrocyte cultures (Table 2). Despite the low number of subjects, the increase in IL-8 reached a statistical trend (p<0.08), and the increase in TNF-α was highly significant (p<0.01).
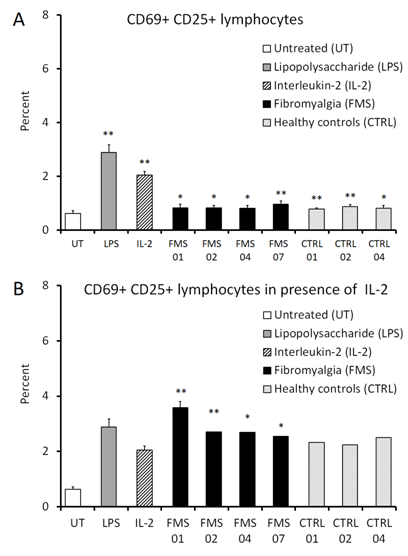
Figure 3 Immune modulation by secreted metabolites from intraerythrocytic microbial forms.
Percentage of CD69+CD25+ double-positive lymphocytes in human peripheral blood mononuclear cell (PBMC) cultures treated for 24 hours with supernatants from 7-day erythrocyte cultures in either the absence (A) or presence (B) of Interleukin-2 (IL-2, 100 IU/mL). Lipopolysaccharide (LPS) was used as a positive control (10ng/mL) for increase in the percentage of CD69+CD25+ double-positive lymphocytes. In the absence of IL-2, 7-day erythrocyte culture supernatants from both fibromyalgia (FMS) patients and healthy controls led to a slight increase in the percentage of CD69+CD25+ double-positive lymphocytes when compared to untreated cultures (A). In the presence of IL-2, only 7-day erythrocyte culture supernatants derived from FMS patient cultures led to an increase in the percentage of CD69+CD25+ double-positive lymphocytes when compared to cultures treated with IL-2 alone (B). Data presented as mean ± standard deviation from quadruplicate cultures using PBMC from a healthy human donor. *P<0.05; **P<0.01.
pg/mL |
IL-2 control |
FMS01 |
FMS02 |
FMS04 |
FMS07 |
CTRL01 |
CTRL02 |
CTRL04 |
p-valuec |
IL-6 |
4.85±1.96 |
71.63±2.00** |
22.28±0.40** |
9.24±0.37** |
5.80±0.18 |
5.18±0.00 |
5.18±0.35 |
6.05±0.18 |
p<0.25 |
IL-8 |
682±245 |
4087±326* |
2788±231* |
2168±64** |
1700±65 ** |
1172±89 ** |
1209±9 ** |
1498±63 ** |
p<0.08 |
TNF-α |
9.85±2.83 |
22.78±2.87 |
17.77±1.4 ** |
17.77±1.40** |
22.77±0.00 ** |
12.89±0.00 * |
8.20±1.30 |
10.99±0.00 |
p<0.01 |
Table 2 Cytokine production by healthy peripheral blood mononuclear cells in response to secreted factors from erythrocyte cultures ab
aSupernatants from erythrocyte cultures were selected based on detection of nucleic acid sequences by PCR The supernatants from fibromyalgia syndrome patients (FMS) and healthy controls (CTRL) were from blood cultures that tested positive for the presence of Aquabacterium rDNA.
bStatistical significance (p<0.05: *, p<0.01: ** respectively) when comparing cytokine levels in cultures treated with erythrocyte culture supernatants to control cultures treated with IL-2.
cStatistical significance is indicated when comparing cytokine levels in cultures treated with erythrocyte culture supernatants from FMS patients to erythrocyte culture supernatants from healthy control subjects.
An underlying, occult microbial involvement has been speculated as a contributing factor for the chronic low-grade inflammation underlying Fibromyalgia syndrome (FMS) and related pathologies such as Chronic fatigue syndrome. The results from the pilot study presented here supports this notion and suggest an advantage for using ex vivo culture of FMS blood samples to study bacterial presence and metabolic activity that may affect the biology of the host. Our methodology showed the amplification of bacterial DNA from the genus Aquabacterium in blood cultures from seven out of eight FMS patients. The data showed increased ability of the FMS erythrocyte culture supernatants, containing bacterial metabolites, to activate immune cells leading to production of inflammatory cytokines. We were also able to amplify Aquabacterium DNA from blood cultures of some healthy controls. However, this was not associated with inflammatory metabolites, suggesting that the bacteria in blood from healthy donors either were less metabolically active, or expressed a different profile of genes.
The choice of cell type for this study is important, since erythrocytes provide many methodological and diagnostic advantages, particularly the lack of nuclei and mitochondria in this cell type, making the use of microscopical observations in combination with fluorescent nucleic acid-binding dyes more attractive and conclusive for bacterial presence. It is documented that certain pathogenic organisms, such as Plasmodium,48 Borrelia [Jensen, unpublished observations], and Bartonella49 can live in intraerythrocytic environments while other bacteria may associate with erythrocytes to avoid immune surveillance.50,51 There is also easy access to iron52 and the potential for intracellular biofilm formation. Previous work has implicated L-form bacteria with disease53,54 as well as shown an association with erythrocytes.55
The Aquabacterium-specific primers resulted in amplification of 16S rRNA sequences in more samples than the 16S universal primers. The Aquabacterium-specific primers, because of their specificity, will be more sensitive in detecting the presence of Aquabacterium DNA than 16S rRNA universal primers. The FMS patient FMS01 was the only individual where freshly isolated erythrocytes gave a positive 16S rDNA amplification product when using Aquabacterium-specific 16S PCR primers. It is possible that this individual had a higher bacterial load in erythrocytes than other individuals or that inhibiting substances present in DNA isolated from erythrocytes could be interfering with the PCR performed with 16S Aquabacterium primers in the other samples. Our data supports the original finding of others where 16S rDNA PCR amplified bacterial DNA from the blood of healthy people56,57 including sequences derived from Aquabacterium.58 In addition, our data takes this finding a step further by suggesting an association with intracellular biofilm formation in erythrocytes. It is known that bacteria in the mouth regularly enter the circulation59 but are presumably detected and neutralized by a properly functioning immune system. In the case of chronic disease, it is possible that a compromised immune system may not be able to respond to this normal influx of bacteria and they may be able to establish an intracellular niche (such as in red or white blood cells) and contribute to chronic inflammation. It is interesting that Aquabacterium is highly abundant in drinking water biofilms47 and biofilms containing bacterial DNA have recently been implicated as the immunogenic agent in the autoimmune disease Systemic Lupus Erythematosus.60
Documentation of microbial presence alone does not provide indication for whether the microbes are metabolically active in a way that may interfere with the host neuroimmune system. Therefore, it was an important part of our pilot study to also evaluate whether the culture supernatants from erythrocyte cultures from healthy donors versus FMS patients showed differences. We found that there were distinct immunogenic properties in the metabolite fraction of the FMS erythrocyte cultures when compared to cultures of healthy erythrocytes. Compounds present in 7-day erythrocyte culture supernatants derived from FMS patients activated immune cells from a healthy donor when PBMC were cultured in the presence of interleukin-2, leading to significantly increased numbers of CD69+ CD25+ lymphocytes. Moreover, in the presence of IL-2 mediated immune activation, the FMS metabolite fraction increased three pro-inflammatory cytokines, IL-6, IL-8, and TNF-α, where the increase in TNF-α reached a high level of significance. This result is noteworthy in the light of mounting evidence for perturbed cytokine profiles in FMS patients and chronic systemic inflammation.61-68 Our results suggest a possible role of metabolically active intraerythrocytic bacteria in contributing to ongoing chronic inflammation, but further research is necessary to solidify the observations in a larger population. The significance of this bacterial presence remains to be determined, and the identification of Aquabacterium does not rule out the presence of other yet unidentified bacterial species. It is possible that additional species of occult bacteria contribute to the metabolite production that was shown to cause increased inflammation in immune cell cultures.
Our findings support the hypothesis that FMS represents a condition of chronic systemic inflammation and suggest that the underlying etiology may involve intraerythrocytic bacteria, present at a higher load than what can be seen in some healthy individuals. In addition, the level of secreted bacterial metabolites appears to differ between FMS and healthy individuals. Microbial presence is not equivocal to illness, but the number of bacteria, combined with their metabolic activity, may be so. Further work is necessary to compare the blood microbiome of FMS patients and healthy controls to monitor fluctuations in bacterial presence and load over time. The observation that the metabolites produced during ex vivo culturing differ between FMS and healthy controls suggest that this may be a useful tool for monitoring bacterial contribution to host inflammatory status, and may also open new avenues to test pharmaceutical and non-pharmaceutical methods to control the bacterial metabolic activity, thus helping to reduce the inflammatory state of the human host.
The study was conducted at NIS Labs. The study was sponsored by the Fetzer Franklin Fund of the John E. Fetzer Memorial Trust.
The authors have no financial interest in the subject matter.

©2019 Benson, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.