International Journal of
eISSN: 2381-1803


Submergence stress regularly affects 15million hectares or more of rainfed lowland rice areas in South and Southeast Asia. These studies will greatly assist in devising more efficient and precise molecular breeding strategies to introgression of SUB1 QTL into the genetic background of HYV rice through marker assisted breeding and to evaluate high yielding rice varieties tolerance to submergence. Seeds of indica rice variety BINAdhan-11 and BRRI dhan52 (submergence tolerance) as a check variety and sub1 introgression lines BPR6, BPR7 were grown in a greenhouse. Genotypes were evaluated in concrete tanks for submergence tolerance under moderate stress. In the present study, molecular markers that were tightly linked with Sub1, flanking Sub1, and unlinked to Sub1 were used to apply foreground, recombinant, and background selection, respectively, in backcrosses between a submergence-tolerant donor and the widely grown recurrent parent. The highest grain yield (5.24t/ha) was obtained from BPR6 followed by BRRI dhan52 (5.03t/ha) and Survival % was also obtained as highest in BPR6 (92%). In case of BPR6, farmers choose this line due to higher yield than others & also attractive grain size. However, it is expected that the promising sub1 line BPR6 will be possible to develop high yielding submergence tolerant variety with three to four weeks tolerance to increase the rice production in the submergence prone areas of Bangladesh where arises single flash flood under different cropping patterns.
Keywords: MAS, extraction, submergence, rice
Rice (Oryza sativa L) is one of the most popular food and feeds over half population of the world. It belongs to the Poaceae family. The existing population of the world is 7.5 billion as of July 2017 and growth rate of around 1.11% per year according to the UNDP approximations elaborated by World meters as a July 2017. The latest United Nations projections indicate that world population will reach 10billion persons in the year 2056 (UNPD, 2015) and rice production must increase by 40-50% to meet the growing demand. In Bangladesh, the current population is 164,957,528 with population density 1266 per Km2 and Bangladesh ranks number 8 by population in the world as of Friday, July 28, 2017, based on the latest United Nations estimates.1 Food scarcity has been and will remain a major concern for Bangladesh as currently (2017) population growth rate is 1.15% per year. But rice and wheat cover 79.4 percent of total cultivable land of Bangladesh, as mentioned in FAO/WFP CFSAM 2008 Report. Global climate change such as floods, drought, cold and saline excesses in precipitation increasingly limit food, fiber, and forest production worldwide.2 In this circumstance, Marker assisted backcrossing (MABC) is one of the key components for breeders that facilitates the use of the results of genomics for farmers’ benefit.3 The applied case of MAS obviously demonstrates the superiority of using MAB compared to conventional backcrossing.4 For this purpose, we need to develop submergence tolerance HYV cultivar and Marker Assistant Selection (MAS) may play a vital role to develop that genotypes.
Flooding of rice fields is a serious problem in the river basins of South and South-East Asia almost in lowland and deep-water rice areas in the world. The estimated annual economic loss of this is more than US$ 600Million. Submergence stress regularly affects 15million hectares or more of rain fed lowland rice areas in South and Southeast Asia.5 However, complete submergence due to flash flood can adversely affect plant growth and yield. In case of Bangladesh situation, rice chronically deficit due to submergence, drought cold and saline. More than 2.0million-hectare areas are affected by different grades of flash floods and reduces 5% average yield in Bangladesh.6 That is why, to develop submergence tolerance rice by using MAS is best option against flash flood.
Rice is one of the most important crucial for the food security of many Asian countries. China and India, the two most populous nations in the world, are also the largest producers and consumers of rice.7 It is grown widely in diverse agro-climatic conditions ranging from high altitudes of the Himalayan hills to the sea coasts of the Indian subcontinent and South-East Asia. Rice is the most extensively cultivated cereal crop in Bangladesh which covers about 74% of the total cropped area.8 In the other words, rice production also has plateaued over recent years through expansion of other crops, other enterprises and development of infrastructure. Traditional varieties and landraces of rice are well adapted to local edaphic factors, including drought, flood, cold, heat, and saline and acid soil conditions. But the static production of rice is still attributable to the lack of suitable improved cultivars for different agro climatic conditions, particularly unfavorable ecosystems.9 The SUB1 QTL on chromosome 9 accounts for 70% of the phenotypic variation for survival under submergence has been fine mapped on chromosome 9 and the cluster of genes underlying the QTL has been cloned.10–12 This QTL has successfully been introgressed into a few different varieties at the International Rice Research Institute (IRRI).5,6,13–15 Moreover, breeding and mutation for enhancement of tolerance to submergence in rice is the major objective in development of varieties for submergence prone, rain fed ecologies. These studies will greatly assist in devising more efficient and precise molecular breeding strategies to introgression of SUB1 QTL into the genetic background of HYV rice through marker assisted breeding and to evaluate high yielding rice varieties tolerance to submergence.
Plant materials
Seeds of indica rice variety BINAdhan-11 and BRRI dhan52 (submergence tolerance) as a check variety and sub1 introgression lines BPR6, BPR7 were grown in a greenhouse (temperature of 30ºC and relative humidity of 75%) at the Biotechnology Division, Bangladesh Institute of Nuclear Agriculture (BINA),Mymensingh, Bangladesh.
Submergence screening
Seedling stage
Seedlings screening by submergence were performed in the submergence tank following standard protocols.11 Seeds were germinated in rows in 20cm X 15 X 10cm trays with three replications. Fourteen-day-old seedlings were submerged for 21days. The survival percentage and elongation ratio of plants were taken 21days after desubmergence. Submergence tolerance score was given just after desubmergence and 7days after recovery following SES of IRRI16 for confirmation of the presence of the Sub1 locus.
The second set of submergence screening was carried out following the same standard protocols.11 Fourteen-day-old seedlings were submerged for 14days. The survival percentage and elongation ratio of plants were taken 14days after desubmergence. Submergence tolerance score was given after desubmergence following SES of IRRI16 for confirmation of the presence of the Sub1 locus.
Vegetative stage
The trails were design by RCBD with three replications in the artificial submergence tank. Genotypes were evaluated in concrete tanks for submergence tolerance under moderate stress. After that Seedling age at 21days were transplanted in the artificial tank and submerged at 30 DAT. After draining of water, the plant height was measured from five randomly selected plants for each entry. Then, the hills could convalesce for 7d and the varieties were recorded visually to categorize into 4 groups: tolerant (scores 1), moderately tolerant (scores 2), moderately susceptible (scores 3) and highly susceptible (scores 4–5), comparing with the check variety (Table 1). Data were taken before submergence and 7d after recovery, and Survival rate: Percent survival was estimated from number of live plants at the 7th day of growth recovery and the total number of seedlings used for a variety.
Score |
% Survival |
Recovery status |
Criteria |
1 |
100 |
Excellent |
High satisfactory growth and tillering , erect dark green leaves |
2 |
90-99 |
Very good |
Satisfactory growth and tillering , erect dark green leaves |
3 |
80-89 |
Good |
Normal satisfactory growth and tillering , erect dark green leaves |
4 |
70-79 |
Fair |
Low growth and tillering , droopy pale green leaves |
5 |
≤60 |
Poor |
Very low growth and tillering , long pale green leaves |
Table 1 Standard Evaluation System(SES) for survival after submergence. Survival data was taken 7 days after submergence. Recovery status after submergence.
Genomic DNA extraction
Leaves sample (3-4 cm-long) from different seedling of each line were collected and it was used for genomic DNA extraction using the mini-preparation protocol17 at Biotechnology Lab., Bangladesh Institute of Nuclear Agriculture (BINA), Mymensingh. Cetyltrimethyl ammonium bromide (CTAB) method is simple and fast compared to other methods and no liquid nitrogen is required.18 The leaf samples were cut into 2-3cm pieces and ground the sample with 670ml extraction buffer and 50ml 20% SDS were added and then mixtures were vortexed for 20 second and incubated for 10minutes at 65°C in the hot water bath. 100ml 5M NaCl was added and inverted gently shaking by hand to suspend the samples evenly. Then added 100ml CTAB and mixed well with vortexed for 20 second and incubated for 10minutes at 65°C in the hot water bath. 900ml chloroform (chloroform: isoamylalcohol=24:1) was added and mixed well. The samples were spinned down at 12000rpm for 15 minutes and then transferred the supernatant into a new Eppendorf tube. Moreover, 600 ml ice-cold isopropanol was added to the supernatant and shaken slowly. Next, the mixture was again Spinned down at 12000rpm for 15minutes by centrifuge. The supernatant was discarded and washed the pellet with 200ml 70% ethanol and then spinned down at 12000rpm for 5minutes. Then the ethanol was removed and the pellets were allowed for air-dried overnight. Eventually pellet was suspended in 100ml 1X TE buffer. Finally, the DNA samples were stored at - 20°C.
Procedure for preparation of PCR cocktail (Every sample)
To begin with, 3.20μl ddH2O, 1.0μl of PCR Buffer (10X), 1.00μl of dNTPs added together in a 0.2ml PCR tube. Then 1μl primer-F and 1μl primer-R were taken in that tube and vortexed. Moreover 0.2μl of Taq DNA polymerase was mixed and finally 2.0μl DNA added for replication.
PCR condition
After initial denaturation for 2minutes at 94°C, each cycle comprises 30 second denaturation at 94°C, 30second annealing at 50-55°C, and 2min extension at 72°C with a final extension for 5minutes at 70°C at the end of 34 cycles.
Adaptability of lines
Two sub1 introgression lines (BPR6, BPR7) along with submergence tolerance standard check variety (BINA dhan-11 and BRRI dhan52) were evaluated at Rangpur region in the northern part of Bangladesh in the real submergence prone environments of the farmers’ field. The experiment was laid out in RCB design with three replications. Thirty days old seedlings of each genotype were transplanted @ 2-3 seedlings/hill with a spacing of 20´20cm. Fertilizer doses were 133:112:75: 60:11 kg/ha Urea, TSP, MP, Gypsum, ZnSO4. All the fertilizers except urea were applied as basal before final land preparation. Urea fertilizer were applied as top dress with 3 equal splits at 15-20DAT, 30-35DAT and 45-50 DAT. Furthermore, pesticide and insecticide were applied according to the crop conditions.
Totally 200 microsatellites or simple sequence repeat (SSR) markers were used for parental survey9 and among this 52 background SSR markers (Table 2). All the SSR markers were selected from the Gramene database (www.gramene.org). For the microsatellite DNA fingerprinting of the eight cultivars such as FR13A, BRRI dhan52, Guti swarna, Mamun swarna, Bilati swarna, Arail, katair, panishail polymorphism was scored according to their molecular weight on polyacrylamide gels. Primers which produced bands at different level were considered as polymorphic marker but those primers which produced bands at the same level were considered as monomorphic. On the other hand, primers which did not produce any bands were considered as not-amplified. Importantly, water was used as negative control for each primer where instead of DNA sample, water was used. This technique is very helpful to detect fake bands produced from any kinds of contamination, or from undesirable nucleotides remaining in the primer or Taq polymerase enzyme.
Chr 1 |
Chr 2 |
Chr 3 |
Chr 4 |
Chr 5 |
Chr 6 |
RM495 |
RM154 |
RM569 |
RM471 |
RM153 |
RM469 |
RM84 |
RM279 |
RM231 |
RM17162 |
RM413 |
RM585 |
RM259 |
RM555 |
RM545 |
RM273 |
RM421 |
RM402 |
RM05 |
RM424 |
RM14963 |
RM17391 |
|
RM275 |
RM237 |
RM341 |
RM411 |
|
|
RM141 |
|
RM208 |
RM570 |
|
|
RM586 |
|
|
RM85 |
|
|
|
Chr 7 |
Chr 8 |
Chr 9 |
Chr 10 |
Chr 11 |
Chr 12 |
RM481 |
RM407 |
SC34 |
RM244 |
RM286 |
RM247 |
RM346 |
RM6208 |
SC16 |
RM25181 |
RM26360 |
RM27877 |
RM336 |
RM72 |
SC26 |
RM25321 |
RM167 |
RM17 |
RM18 |
RM256 |
SC30 |
RM258 |
RM26482 |
|
|
|
RM242 |
|
RM206 |
|
|
|
RM278 |
|
RM254 |
|
Table 2 List of initial background markers of all 12 chromosomes used in the background selection
The microsatellite enriched DNA fingerprints were constructed using the standard procedures. In our study, primers tightly linked to sub1 genes, flanking sub1 genes for selection. Moreover, application of background primers for the selected plants were used here (Table 3). In this study, two sub1 introgression lines BPR6, BPR7 of rice were evaluated using 3 primer pairs (RM 18, RM586, and RM17391). These three primer pairs were used to evaluate sub1 introgression lines BPR6, BPR7 with donor parent BRRI dhan52 by amplification of the DNA sequence to submergence tolerance. The bands obtained BPR6, BPR7 lines were compared to the band obtained from submergence tolerance BRRI dhan52. The BPR6, BPR7 lines having similar banding pattern to BRRI dhan52 were considered as submergence tolerant (Figure 1). Confirmation of sub1 gene in the selected plants were tested with the cleaved amplified polymorphic sequence (CAPS) marker specific to sub1 gene which was the putative candidate marker for sub1 gene. The tested plants showed the same banding pattern of resistant allele of the donor of sub1 gene BRRI dhan52. So, it was possible to confirm that submergence tolerant (sub1 gene) has been introduced to these lines. Among the several primers, the established SSR markers of this study provided a positive assessment in making unique DNA profiles and identity of the genotypes. The selected submergence tolerant rice lines would be further tested in flood prone areas to observed yield potentiality for developing high yielding submergence tolerant varieties. The present study has the potential applications in future breeding programme for the genetic improvement of submergence tolerant rice.
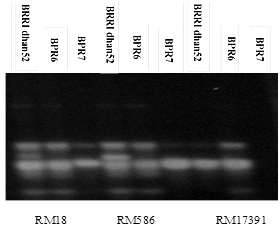
Figure 1 Confirmation of different alleles in the sub1 introgression lines BPR6, BPR7 rom donor parent BRRI dhan52 of RM 18, RM586, and RM17391 for Background selection.
Primer |
Position (bp) |
Chro. locus |
Sequence |
Ann. Temp. |
Repeat motif |
|
Forward |
Reverse |
|||||
RM18 |
25,652,511-25,652,668 bp |
7 |
5’-TTCCCTCTCATGAGCTCCAT-3’ |
3’-GAGTGCCTGGCGCTGTAC-5’ |
55 |
(GA)4AA(GA)(AG)16 |
RM586 |
1,476,793-1,477,087 bp |
6 |
5’-ACCTCGCGTTATTAGGTACCC-3’ |
3’-GAGATACGCCAACGAGATACC-5’, |
55 |
(CT)23 |
RM17391 |
29,279,887-29,280,062 bp |
4 |
5’-ACTTTGCTCTGAACTTGCAGTGG |
3’GCTAGCTGCTATCAGGATTCACG-5’. |
55 |
(AG)23 |
Table 3 The sequence and position of background markers of three chromosomes used in the background selection.
The Rangpur regions at northern zone are flood prone area of Bangladesh in Aman season. An experiment of validation trials was conducted to evaluate with the performance of yield and yield contributing characters at Rangpur region. There was two times. There was 3days of flash flood during active tillering stage in this trial with the average water height of 85cm (Figure 2). Second flash flood was 3days with the average water height of 90cm (Table 4). The genetic variability among Aman rice genotypes such as BPR6, BPR7 (two lines), BINAdhan-11 and BRRI dhan52 (two varieties) to demonstrate of duration, plant height, filled grain, unfilled grain, number of panicle,1000-grain wt, survival rate and yield. The average growth duration of BPR6, BPR7 BINAdhan-11 and BRRI dhan52 were 142,145,123,142days with the average yield of 5.24t/ha, 4.76t/ha, 4.72t/ha & 5.03t/ha respectively. BPR6 line produced the highest number of panicle hill-1 (10) than other varieties (9). The highest mean (122) for number of filled grains panicle-1 was found in BPR6 and the lowest (118) in BRRI dhan52. The highest mean value (25) for number of unfilled spikelets panicle-1 was observed in BPR6, BPR7 and BRRI dhan52 but the lowest (22) in BINAdhan-11. BPR6 and BRRI dhan52 produced the heaviest (27.0g) grains whereas that of the lightest (22.6g) grains was observed in the BPR7.The highest grain yield (5.24t/ha) was obtained from BPR6 followed by BRRI dhan52 (5.03t/ha). Survival % was also obtained as highest in BPR6 (92%) in Table 5. Among the genotypes the farmers choose BPR6 due to tall plant type & for better yield because of average plant height was 116cm and it was more straw for cattle. In case of BPR6, farmers choose this line due to higher yield than others & also attractive grain size. From the farmers view, evaluated BPR6 to grow for increasing total productivity to insure food security.
Flooding status |
Duration |
Days |
Water height |
First Flood |
15-17 August,2015 |
3Days |
85cm |
2nd Flood |
26-28 August,2015 |
3Days |
90cm |
Table 4 Flood status
SN |
Genotypes |
Plant height |
Growth duration |
Panicle/Hill |
Total grain/panicle |
1000 grain wt(gm) |
Grain Yield |
Survival (%) |
|
Filled grain |
Unfilled grain |
||||||||
1 |
BPR6 |
116 |
142 |
10 |
122 |
25 |
27 |
5.24 |
92 |
2 |
BPR7 |
91 |
145 |
9 |
122 |
25 |
22.6 |
4.76 |
91 |
3 |
BINAdhan-11 |
108 |
123 |
9 |
121 |
22 |
23.8 |
4.72 |
88 |
4 |
BRRI dhan52 |
116 |
142 |
9 |
118 |
25 |
27 |
5.03 |
91 |
Table 5 Average performance of the entries under validation trials in Rangpur region, T. Aman 2015
The results presented here that the utilization of marker assistance selection (MAS) technique for developing the submergence tolerance lines from the parent mega variety. Iftekharuddaula et al.,19 reported that, submergence tolerance variety as a high yield was developed by using BR11 as recipient parent applying foreground, phenotypic and background selection approaches. To minimize linkage, drag recombinant selection were most essential by backcross populations. The BRL were developed higher yield with 14days complete submergence compared to the NIL populations at BRRI. The Backcross Recombinant Line IR85260-66-654-Gaz2 was released as BRRI dhan52 in 2010 which was the first high yielding submergence tolerant variety and average yielding ranging 4.2-5.2t/ha in Bangladesh. This fingerprinting and genomic relations of this cultivars will be useful for Bangladesh breeder for developing new high yielding verities. The higher submergence tolerance of the BRL IR85260-66-654-Gaz2 (BRRI dhan52) might be associated with some minor genes remaining in the 15 Mb donor segment on the carrier chromosome which again can be inherited from FR13A in the donor parent IR40931-33-1-3-2 or might be due to some positive interaction of SUB1 QTL with the genes remaining in that region. Submergence tolerance is a polygenic trait, and SUB1 QTL does not completely represent the trait alone.20 The donor parent of the present study is already a moderately improved breeding line21 which possesses very strong submergence tolerance. The introgression size in the three BRLs viz. IR85260-66-217, IR85260-66-258, IR85260-66-654 was 15 Mb on chromosome 9 including SUB1 QTL from IR40931-33-1-3-2. In conventional backcrossing, large donor segments are likely on the carrier chromosome.22 Neeraja et al.,5 utilized a major QTL on chromosome 9 called sub1, to apply marker assisted backcrossing (MAB) for the development of submergence tolerant versions of rice cultivars that were widely grown South Asia. By using the BC2F2 generations a submergence tolerance rice was obsessed Swarna type SSRs alleles analyzed except tip segment of chromosome 9 for sub1 locus. A BC3F2 double recombinant plant was identified that was homozygous for all Swarna type alleles except for an approximately 2.3-3.4 Mb region surrounding the sub1 locus. It follows that, mega variety Swarna could be proficiently transformed to a submergence tolerance variety by using three years backcrossing. Polymorphic markers for foreground and recombinant selection were identified for four other mega varieties to develop a wider range of submergence tolerant varieties to meet the needs of farmers in the flood-prone regions. This method indicates the current use of marker assistance selection (MAS) for major QTL in molecular sciences. Moreover, BRRI dhan52 was the donor parents of this “second generation” of sub1 line that hold high yield and high submergence tolerance with agronomic traits. In conventional backcrossing, large donor segments are likely to be introgressed in the carrier chromosome22 and, in this approach, a large donor segment has been inherited from the carrier chromosome due to the lack of recombinant selection done in early backcross generations. Young & Tanksley22 also pointed out that the donor genes on the carrier chromosome were the most difficult to eliminate, and could persist long after the donor genome content of non-carrier chromosomes has returned to approximately zero if no selection using markers was applied. Since the donor parent was an improved genotype, this large introgression did not cause any negative effect on the phenotype of BRRI dhan33-Sub1 lines. The performance of eight entries including BRRI dhan51 and BRRI dhan52 presented that lowest duration from BRRI dhan52(148days) and plant height ranged 94-120cm through one weak complete submergence. The highest grain yield (4.85t/ha) was obtained from BRRI dhan52 followed by from BRRI dhan51 (4.6t/ha). More than 90% survival obtained from all the entries indicated that there was nor much stress of flash flood in this trial.23 BRRI dhan52 as a submergence tolerance variety with high yielding showed 14days tolerance against flash flood of Bangladesh. But some parts of Bangladesh such as northern part occur 21 to 28days duration of flash flood. That is why, it was expected that the promising sub1 line BPR6 will be possible to develop high yielding submergence tolerant variety with three to four weeks tolerance to increase the rice production in the submergence prone areas of Bangladesh where arises single flash flood under different cropping patterns.
There was almost 2m ha land area affected by different grades of flash floods and reduces 5% average yield in Bangladesh. The present study was therefore undertaken to introgression of SUB1 QTL into the genetic background of HYV rice through marker assisted breeding and to evaluate high yielding rice varieties tolerance to submergence. However, it is expected that the promising sub1 line BPR6 will be possible to develop high yielding submergence tolerant variety with three to four weeks tolerance to increase the rice production in the submergence prone areas of Bangladesh where arises single flash flood under different cropping patterns. Further research is needed, however, before the use of such materials can be recommended for all farmers in Bangladesh.
My foremost acclamation, praises and gratitude is to God almighty and deepest recognition and very special thanks are due to my colleagues in the workplace who were there to help me in every moment of this huge work. Chemicals and instruments are financial supported by BINA.
The author declares no conflict of interest.

© . This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.