eISSN: 2373-6372


Research Article Volume 9 Issue 6
1School of Life Sciences, Jawaharlal Nehru University, India
2Department of Gastroenterology, All India Institute of Medical Sciences, India
Correspondence: Dr. Jaishree Paul, School of Life Sciences, Jawaharlal Nehru University, New Delhi, 110067, India
Received: May 15, 2018 | Published: December 31, 2018
Citation: Ranjha R, Aggarwal S, Ahuja V, et al. Site specific expression of CXCL-12 ? and its regulation by miR-200a in ulcerative colitis. Gastroenterol Hepatol Open Access. 2018;9(6):290-295. DOI: 10.15406/ghoa.2018.09.00345
Purpose: CXCL-12β is a chemokine playing an important role in inflammation and cancer. Altered cytokine and chemokine expression is a hallmark of ulcerative colitis. Emerging evidence suggest that microRNAs are involved in inflammatory process and cancer development during chronic inflammation. This study was done to find the regulation of CXCL-12 expression via different miRNAs in UC.
Methods: Biopsy samples were collected from UC patients and non-IBD controls. CXCL-12β expression was analyzed at mRNA as well as protein level in UC and compared to controls. Expression of miRNAs targeting CXCL-12β was analyzed by microarray and real time PCR. In vitro studies were done to validate CXCL-12β as a target of miR-200a-3p.
Results: Expression of CXCL-12β was increased in UC compared to controls. MiR-141-3p and miR-200a-3p were predicted to target CXCL-12β. Expression of miR-141-3p was upregulated and expression of miR-200a was downregulated in UC compared to controls. In vitro studies using reporter assay proved CXCL-12β as a target miR-200a. Expression of CXCL-12β increased while expression of mir-200a decreased in the cancer prone, rectosigmoid area of colon.
Conclusion: Our study reveals that miR-200a-3p regulates the expression of CXCL-12β in UC. Both CXCL-12β and miR-200a also show site specific altered expression in the colon of UC patients. Our data shows that regulation of CXCL-12β expression using miRNA may be a promising therapeutic approach in UC pathogenesis.
Keywords: Ulcerative Colitis, Cancer, MiRNA, CXCL12
IBD is a chronic inflammatory disease of gastrointestinal tract which involve complex interactions between immune system, genetic makeup, gut microbiota and environmental factors.1 Ulcerative Colitis (UC) is one of the two main subtypes of Inflammatory Bowel Disease (IBD).2 Along with other pathogenic factors, altered cytokine and chemokine expression play important role in the pathogenesis of UC.3,4 Patients with prolonged UC have increased risk of developing colorectal cancer (CRC).5–8
Expression of pro-inflammatory cytokines and chemokines are known to increase in UC.3,9The degree of inflammation and tissue damage is dependent on the concentration of local chemokines. Thus over expression of chemokines leads to enhanced recruitment of immune cells to the inflammatory site.9 Experimental evidences show that CXCR4/CXCR7/CXCL12 chemokine axis play crucial role in inflammation as well as cancer. CXCL12/CXCR4 chemokine axis is known to be involved in pathogenesis of IBD specially UC.10 CXCL12 is expressed by normal intestinal epithelial cells (IECs) and upregulated in inflammed IECs of IBD patients.11 Interestingly, enhanced expression of CXCL-12 was observed in UC compared to CD and more specifically in inflammed compared to non-inflammed mucosa.12 Upregulation of CXCL12 in the inflammed colonic mucosa is specifically due to the variant CXCL12β upregulation.13 Polymorphisms in CXCL12 are also reported as risk factor for IBD development.14,15 Blockade of CXCL12/CXCR4 axis has been shown to ameliorate colitis in both DSS induced as well as in IL-10 KO mice model of colitis. Antagonist of CXCR4 also showed potent anti-inflammatory effect.10
Micro RNAs (miRNA) are small non-coding RNA molecules that are emerging key players in pathology of many diseases including IBD and cancer.16,17 MiRNA are differentially expressed in colonic mucosa and serum of IBD patients and regulate important genes involved in the disease pathogenesis.11,18,19 It has been shown that miRNA plays important role in fine-tuning of TLR signaling and maintaining homeostasis of gut microbiota, two very important players in the pathology of IBD. 20,21
There is increased risk of cancer development due to chronic inflammation.22,23 There is also an enhanced risk of CRC development in IBD patients specially UC.3 In UC, cancer development predominantly occurs in the rectosigmoid part of colon.6,25 MiR-141 regulate the expression of CXCL-12β , which further mediate leukocyte migration in murine model of colitis and CD.13 MiRNAs are reported as connecting link between inflammation and cancer.22,23,26 CXCL12 is a pro-angiogenic chemokine that act as important player in chronic inflammation as well as cancer development.27,28 Different miRNAs are reported to regulate the expression of CXCL12 in various cancers.13,27,29–39 Earlier studies have reported that tumor promoting miRNAs are overexpressed and tumor suppressing miRNAs are dowregulated in cancer prone rectosigmoid part of colon in UC.4
In the present study we investigated the regulation of CXCL12β through miRNA in UC. CXCL12β expression was determined in colon biopsy samples as well as serum samples of UC patients and controls. The impact of miR-200a over-expression on level of CXCL12β was determined.
Patient Samples and cell lines: 30 UC patients and 25 non-IBD controls were included in this study. 20 samples from UC patients were from rectosigmoid region of colon and 10 were from ascending region of colon. Control samples were non IBD individuals without any IBD symptoms and without any colon inflammation. All control and patient samples were age and sex matched. Diagnosis of UC patients were based on the clinical findings based on ECCO guidelines.5 UC was confirmed by histopathological analysis of colon biopsy samples. This study was approved by Institutional Ethics Review Board, JNU (REF no. 2012/student/28) and the ethical committee of All India Institute of Medical Sciences, New Delhi, India (Ref no. IEC/NP-320/2012). A written consent was taken from all the participants included in this study. Human colorectal adenocarcinoma cell line, HT-29 (National Center for Cell Sciences, Pune, India) were cultured in Dulbecco's Modified Eagle's medium (Gibco, Thermo Fisher Scientific, USA) for in vitro studies.
RNA isolation and RNA quality check: mirVana miRNA isolation kit (Ambion INC, TX 78744, USA) was used for total RNA isolation. RNA was stored at -80°C until further analysis. RNA quality was assessed by running it on 1.5% agarose gel for evaluating the three bands 28S, 18S and 5S rRNA. RIN value for the total RNA was calculated using bioanalyser (Agilent Techonologies, California, USA). Samples with RIN value more than 7 were included for microarray analysis.
Microarray Analysis: MiRNA microarray was performed by Exiqon A/S, Denmark via miRCURY LNA™ microRNA Array (7th gen). Five RNA samples were pooled in each category in equal concentration.6 The array data has been submitted in NCBI Gene Expression Omnibus (GEO) under the entry series GSE66932.
MiRNA target prediction and qRT-PCR: Target prediction for miRNAs targeting 3’UTR of CXCL12β was done using online target prediction tools ( Targetscan, Diana-microT and PicTar). RNA was reverse transcribed to cDNA using miRNA specific stem loop primers for miRNAs7 and Random Hexamer primers were used for cDNA synthesis for other genes by revert aid cDNA synthesis kit (Fermentas St. Leon Rot, Germany). Table 1 shows the primers sequences used for reverse transcription and real time PCR analysis. Real time PCR was performed using ABI PRISM 7500 Real time PCR system (Applied Biosystems). U6 snRNA and GAPDH were used as internal controls. Parameters for real time were- initial denaturation- 94°C for 2 min, denaturation- 94°C for 30 sec, annealing- 60°C for 1 min for 40 cycles. 20 μl creactionontaining 7 μl MQ water, 10 μl SYBER Green universal PCR Master Mix (Applied Biosystems, California, USA), 1 μl of each forward and reverse primer (4 picomole/μl), 1 ul cDNA. The relative expression was calculated using the equation 2-ΔΔCT where ΔΔCT = ΔCT target -ΔCT control.
Name |
Primer (5′-3′) |
|
Reverse Universal Primer for miRNA |
GTGCAGGGTCCGAGGT |
|
hsa-miR-141-3p |
RT Primer |
GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGCCATCTTT |
Forward |
TAACACTGTCTGGTAAAGAT |
|
hsa-miR-200a-3p |
RT Primer |
GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACATCGTT |
Forward |
TAACACTGTCTGGTAACGAT |
|
CXCL-12β |
Forward |
CCTGCCACAGCCTCCCCT |
Reverse |
TAAGCTGCTACGTGTCGCC |
|
Table 1 Sequence of primers used in Reverse Transcription and qRTPCR.*
* RT primers were used for cDNA synthesis in each case; Reverse universal and forward primers were used for real time analysis for each miRNA
Cloning and transfection: miR-200a was cloned in pBABE-puro vector. DNA fragment containing sequence of miR-200a and its flanking region was used for cloning. Primers used for miR-200a cloning were Forward miR-200a 5’- CGCGGATCCACAGCCATCTTCCCTCCTG- 3’ and Reverse miR-200a 5’- CCGGAATTCGGAGCTGACACAGGCCCTC- 3’.8 3’UTR of CXCL12β was cloned in pMIR-Report vector downstream of luciferase gene. Primers used for cloning were Forward CXCL12β 5’-CGAGCTCGGGTCAGACGCCTGAGGAAC-3’ and Reverse CXCL12β 5’ –CGACGCGTACATTTGAGTACAGAACTTTATT- 3’. Transfection of plasmids was done using Lipofectamine 3000 (Thermofisher Scientific, USA) following manufacturer’s protocol.
Enzyme Linked Immunosorbant Assay (ELISA): CXCL-12β concentration in the patient serum samples and cell culture supernatant was measured using ELISA kit (Sunred Biological, Shanghai). The assay was performed according to manufacturer’s protocol and data was analysed using curve expert 1.4 software.
Luciferase assay: Luciferase assay was performed using Dual luciferase assay kit (Promega, USA). PRL-TK vector was used for normalization. CXCL12β 3’UTR containing luciferase vector was transfected in HT-29 cells with or without miR-200a overexpression.
Statistical analysis: Statistical analysis was done using two-way student’s t test. Data analysis was done using Graphpad Prism 5.0. A probability level of p < 0.05 was considered statistically significant.
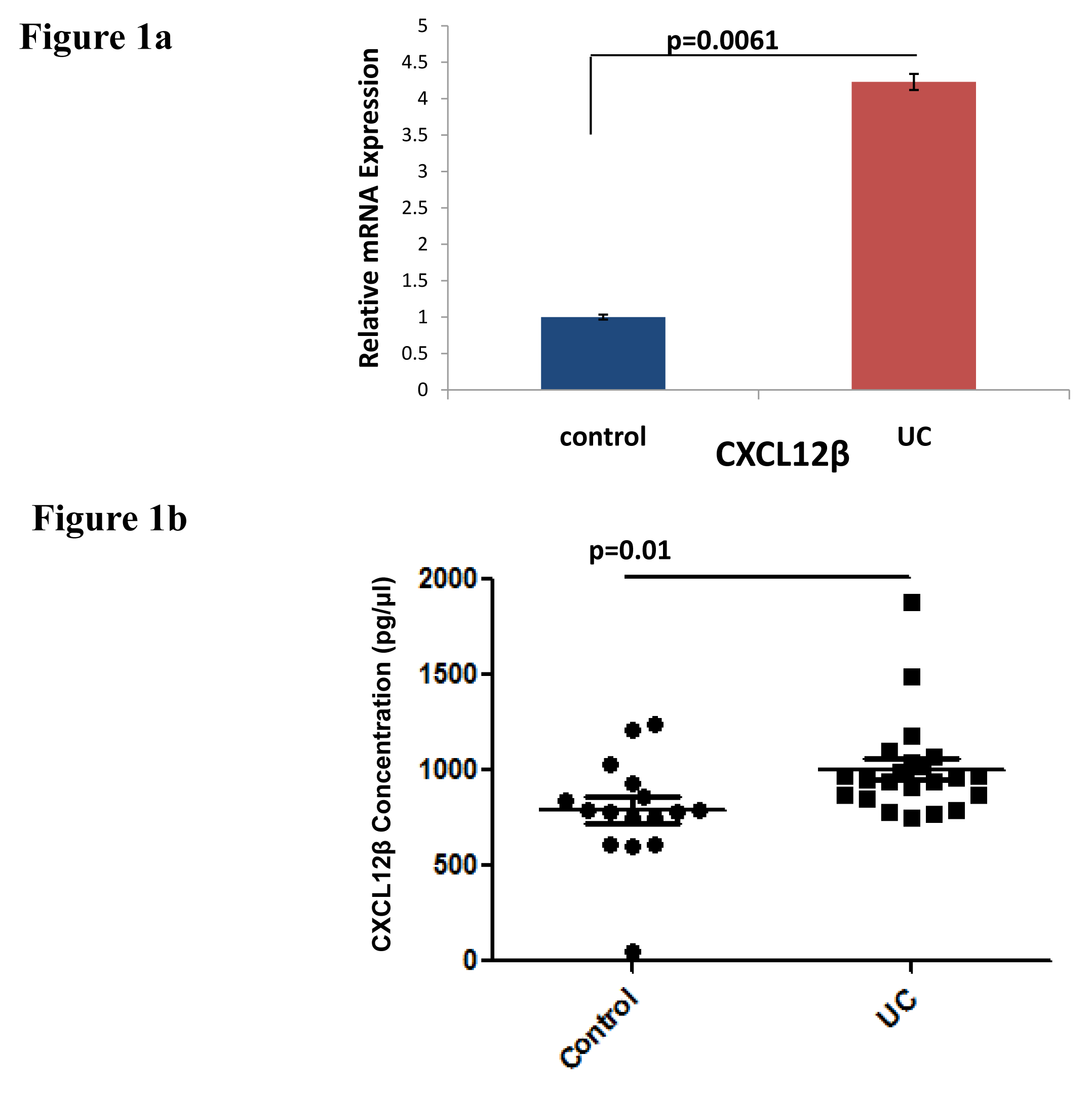
CXCL12β expression in UC vs Control individuals: CXCL12β expression was analyzed both at mRNA as well as protein level. There was significant increase in CXCL12β expression in biopsy samples from UC patients compared to controls (Figure 1a). Level of CXCL12β was increased in the serum samples of UC patients compared to non IBD controls (Figure 1b).
MiRNAs targeting 3’UTR of CXCL12β: Target prediction analysis showed miR-141-3p and miR-200a, members of miR-200 family, targeting 3’UTR of CXCL12β. Both target same two sites, position 51-58 and 2940-2948 in the 3’UTR of CXCL12β (http://www.targetscan.org/cgi-bin/targetscan/vert_71/view_gene.cgi?rs=ENST00000374429.2&taxid=9606&members= &showcnc=0&shownc=0&showncf1=&showncf2=&subset=1).
Expression of miR-141-3p and miR-200a: miRNAs expression was analysed in UC compared to non-IBD controls using microarray analysis. Figure 2 represents the heatmap for the microarray data describing the expression profile of miRNAs in UC and non-IBD controls. Expression of both the miRNAs was decreased in UC compared to control. Microarray data was validated using qRTPCR. MiR-141-3p showed upregulation in our qRTPCR analysis in contrary to microarray data (Figure 3a). Expression of miR-200a however decreased significantly in UC patients compared to controls in qRTPCR results similar to microarray data (Figure 3b).


In vitro validation of CXCL12β as a target of miR-200a: In vitro validation was carried out by miR-200a overexpression and reporter assay.
Effect of miR-200a overexpression on CXCL12β secretion: We looked at the effect of miR-200a overexpression on CXCL12β secretion in HT-29 cell line. Secretion of CXCL12β decreased in the supernatant of HT-29 cells overexpressing miR-200a compared to control transfected and non-transfected cells (Figure 4).
Dual Luciferase assay: Further validation of CXCL12β as miR-200a target was done using 3’UTR reporter gene assay. Luciferase was cloned upstream of 3’UTR of CXCL12β and was used as a reporter. So the luciferase activity was controlled by miR-200a. The level of luciferase activity was decreased by overexpression of miR-200a (Figure 5).


Site specific expression of miR-200a and CXCL12β in colon of UC patients: We looked at the site specific expression of CXCL12β and miR-200a in rectosigmoid vs ascending colon of UC patients. Expression of miR-200a and CXCL12β showed contrasting pattern of expression. Expression of CXCL12β was increased in rectosigmoid colon in UC patients compared to controls. Expression of the chemokine in ascending colon was comparable to non-IBD controls (Figure 6a). Expression of miR-200a was decreased in rectosigmoid colon compared to ascending colon of UC patients and to non-IBD controls (Figure 6b).
CXCL12β is reported to play important role in chronic inflammation and various type of cancers.12,27,28 Different miRNAs were reported to regulate CXCL12 expression in different diseases.13,27,29–39 In this study we investigated how the expression of CXCL12β is regulated by miRNA during UC. We also looked at site specific expression of CXCL12β and its targeting miRNA in context to CRC susceptibility of various parts of colon.
CXCL12 is a constitutive cytokine reported to play important role in various inflammatory diseases including lung, brain, joint and intestinal inflammation. CXCL12 is constitutively expressed by colon epithelial cells. Its expression was reported to be increased in IBD. Its expression is more pronounced in UC compared to CD and in inflamed mucosa compared to non-inflamed mucosa. CXCL12-CXCR4 axis contributes to intestinal epithelial cells migration, barrier function, restitution and cAMP mediated cellular functions in normal intestinal mucosa.44,45 Increased CXCL12 expression attracts immune cells which infiltrate and secrete proinflammatory cytokines in inflamed mucosa.13 CXCL-12 has different splice variants. Due to four additional residues at its C-terminus, CXCL-12β variant is found more resistant to proteolysis. It can maintain a more potent concentration in the blood in order to recruit more leukocytes.46,47 Increased expression of CXCL12 in colonic mucosa is mainly contributed by increased expression of CXCL12β.13 We looked at the expression of CXCL12β in UC vs non-IBD controls. We also found increased expression of CXCL12β at mRNA level in colonic mucosa (Figure 1a) and at protein level in the serum (Figure 1b) of UC patients compared to non-IBD controls.
We looked for the miRNAs targeting 3’UTR of CXCL12β. It was predicted to be target of miR-141-3p and miR-200a. Both are member of miR-200 family and target same site in the 3’UTR of CXCL12β. Expression of both miR-141-3p and miR-200a was decreased in our microarray analysis (Figure 2). We looked at the miR-141-3p expression in the colon of UC patients and controls. MiR-141-3p was reported to regulateCXCL12β expression in the colon of CD patients.13 Expression of miR-141-3p was increased in the colon of UC patients compared to non-IBD controls in our real time analysis (Figure 3a). As miR-141-3p showed same expression as that of target gene, it revealed that probably miR-141-3p was not regulating the expression of CXCL-12β in case of UC. Our real time expression data was in disagreement with our microarray data. Microarray data was often reported to show disagreement with real time analysis.9 We next did real time PCR to validate the expression of miR-200a-3p. Real time expression analysis showed that there was a significant decrease in the expression of miR-200a-3p (Figure 3b), as found in microarray analysis. This indicated that miR-200a-3p might be regulating the expression of CXCL-12β in UC.
MiR-200a-3p was transfected and overexpressed in HT-29 cell line. Increase in expression of miR-200a-3p resulted in significant decrease in secretion of CXCL-12β from HT-29 cell line (Figure 4). We performed dual luciferase assay in order to confirm if the regulation of CXCL-12β is mediated by miR-200a-3p. Increase in expression of miR-200a-3p leads to significant decrease in luciferase activity (Figure 5).
During the process of intestinal inflammation, increased level of CXCl-12β attract different immune cells, which further secrete a large volume of pro-inflammatory cytokines. CXCL12 is a constitutive and inflammatory chemokine in the intestinal immune system. Selective manipulation of the CXCL12/CXCR4 axis had been suggested as potential therapeutic mechanism in IBD.13 Blockade of CXCL12/CXCR4 Axis ameliorates murine colitis in experimental mice model of colitis.10 It has been hypothesized that the CXCL12β targeted by miR-141 in colonic epithelial cells serves as an important mechanism regulating colonic CXCL12β/total-CXCL12 expression and control immune cell trafficking during both experimental colitis models and the CD dysfunction process in humans. Our expression data in patients and validation by in vitro studies proves our hypothesis that the expression of miR-200a-3p regulates the level of CXCL-12β chemokine in the colonic cells during ulcerative colitis.
Rectosigmoid part of the colon of UC patients was reported to be cancer prone in inflammation associated carcinoma development. MiRNA expression was also altered in the rectosigmoid part of colon compared to ascending colon in UC.4 Proinflammatory CXCL12-CXCR4 signaling axis has been reported to play important role in cancer development.10 We looked at the site specific expression of CXCL12β and its targeting miRNA. There was significant increase in expression of CXCL12β and the expression of miR-200a decreased significantly in the rectosigmoid part of colon compared to ascending colon and non-IBD controls (Figure 6a and 6b). This further strengthen our analysis that miR-200a regulate the expression of CXCL12β in UC and do so in a site specific manner. It also showed that CXCL12 may be an important factor in UC associated CRC development as it exhibited increased expression in the cancer prone rectosigmoid part of colon.
In conclusion our study show regulation of CXCL-12β in UC is mediated by miR-200a, not by miR-141-3p as in CD. Our results further revealed that miR-200a was predominantly decreased in the colonic epithelial cells of UC patients leading to increase in expression of CXCL-12β in the colon. Both CXCL12β and its targeting miRNA showed site specific expression in UC. Inhibiting CXCL-12β expression using miRNA may prove to be a valuable approach for treatment of IBD and to prevent IBD associated cancer development.
The current work was financially supported by the Department of Biotechnology, New Delhi DBT grant (award no. BT/PR8348/MED/30/1023/2013 awarded to JP. We acknowledge umbrella funding from DST PURSE II : File No. 6(54)/SLS/JP/DST PURSE/2016-17 awarded to JP. We thank the participants who contributed samples for this study. RR gratefully acknowledges the research fellowship from Council of Scientific and Industrial Research, New Delhi.
The authors declare that no conflicts of interest.

©2018 Ranjha, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.