eISSN: 2373-6372


Research Article Volume 7 Issue 4
1Department of Internal Medicine, Gastroenterology, Hepatology and Endoscopy Unit, Mansoura University, Egypt
2Department of Histology and Cell Biology, Faculty of Medicine, Mansoura University, Egypt
3Department of Microbiology, Al-Azhar University, Egypt
Correspondence: Mohammed Amin Mohammed, Assistant Professor of Internal Medicine, Gastroenterology, Hepatology and Endoscopy Unit, Faculty of Medicine, Mansoura University, Mansoura, Ahmad Maher Street, 6th October Street, Postal Zip code: 35511; Egypt, Tel 00201122022900; 00201001409982; 0020502941497
Received: July 14, 2017 | Published: September 1, 2017
Citation: Mohammed MA, Omar NM, Mohammed SA, Deiab AG (2017) The Significance of Vitamin D Receptor Gene Polymorphisms for Susceptibility to Hepatocellular Carcinoma in Subjects Infected with Hepatitis C Virus. Gastroenterol Hepatol Open Access 7(4): 00246. DOI: 10.15406/ghoa.2017.07.00246
Background: Vitamin D has emerging roles in fibrogenesis, cell cycle arrest, immune modulation, and tumorigenesis. Several single nucleotide polymorphisms (SNPs) in vitamin D receptor (VDR) gene are associated with tumorigenesis in various organs.
Objective: to investigate the association between the VDR gene polymorphisms and Hepatocellular carcinoma (HCC) risk and severity in Egyptian patients with chronic hepatitis C (CHC).
Methods: Five hundred thirty outpatients with chronic hepatitis C virus (HCV) infection were initially enrolled, of which 180 patients with CHC, 180 patients with liver cirrhosis and 170 patients with HCC. Another 170 age- and sex-matched healthy controls were enrolled. Genotyping of VDR gene at BsmI, ApaI and TaqI loci was performed. Evaluation of clinicopathological features of HCC was done. Data were analyzed using SPSS software (Version 17.0).
Results: HCC patients had significantly higher frequencies of ApaI CC genotype and bAt[CCA]-haplotype than non-HCC patients. Patients carrying ApaI C allele, ApaI CC genotype or bAt[CCA]-haplotype had significantly higher prevalence of HCC, advanced stage of liver cirrhosis and lower serum 25-hydroxy vitamin D3 concentrations. Also, after adjusting other covariates (age, gender, platelets count, and serum ∞-fetoprotein and vitamin D levels), patients carrying ApaI CC genotype or bAt[CCA]-haplotype had significantly higher risk for HCC development.
Conclusion: The present study provided significant associations of VDR gene SNPs (ApaI CC genotype and bAt.CCA-haplotype) with HCC development and disease severity in HCV-infected Egyptian patients. Thus, the determination of these VDR genetic variants in HCV patients could help identification of patients at-risk of HCC development.
Keywords: Vitamin D receptor gene; bAt[CCA]-Haplotype; single nucleotide polymorphisms; chronic hepatitis C; hepatocellular carcinoma
Vitamin D has pivotal immunomodulatory actions and emerging roles in hepatic pathophysiology, inflammatory and metabolic liver diseases. It is considered as a systemic hormone that is involved in metabolism, immune regulation, fibrogenesis and cancer development through its actions on Vitamin D receptor (VDR), an intracellular nuclear hormone receptor; subfamily 1, group1, member 1 (NR111) which interacts with target genes producing a variety of biological conformational changes.1 VDR gene is located on chromosome 12q13.11; highly polymorphic contains eleven exons and spans about 75kb.2 Also, it has been previously well established that vitamin D exerts an array of tissue and cell specific antitumor activities.e.g. cell cycle arrest, cell adhesions, inhibition of cancer cell invasion, anti-proliferation, pro-apoptosis, anti-angiogenesis, pro-differentiation on many cancer cells expressing VDR.3 Vitamin D-binding protein (DBP) and 25-hydroxy-vitamin D3 (25-OHD3) are mainly synthesized in the liver. Vitamin D has many direct effects on the liver (e.g. anti-fibrotic and expression of detoxifying enzymes). An association has been established between low levels of vitamin D3 and several diseases including viral hepatitis and cancer development. Low serum levels of 25-OHD3 were proved to be associated with increased liver fibrosis in patients with non-alcoholic steatohepatitis4 and type 2 diabetes mellitus5 and reported to be associated with high risk of HCC development.6 Moreover, the serum level of 25-OHD3 had been reported to affect the natural course and treatment response of chronic hepatitis C.7 Higher vitamin D concentrations were proved to be associated with better prognosis and improved outcomes of adverse health problems.8 Despite the reported mechanisms supporting the beneficial effects of Vitamin D3 supplementation, the total benefits of its supplementation remain ambiguous.
Chronic HCV infection is a major health problem worldwide that is complicated by liver cirrhosis and ultimately by HCC.9 Hepatocellular carcinoma is one of the most common and highly malignant cancers worldwide with high incidence in Sub-Saharan Africa and Eastern Asia.10 The carcinogenesis of HCC is a complex, multifactorial and multistep process. HCC has a very bad prognosis and restricted treatment options. There are multiple risk factors which contribute to hepatic carcinogenesis and HCC development including chronic hepatitis C (CHC) or chronic hepatitis B (CHB), liver cirrhosis, carcinogen exposure (dietary aflatoxin), and chronic alcohol ingestion.11
Single-nucleotide polymorphisms (SNPs) of VDR gene may deregulate the activity of vitamin D, affecting the cancer risk. It has been reported that SNPs in the VDR gene are associated with tumorigenesis in various organs such as prostate,12 breast,13 skin,14 colon and rectum,15 and kidneys16 but these observations are conflicting and debatable.17 The interaction between VDR gene polymorphisms and vitamin D status has a pivotal role in cancer prognosis.18 Previous epidemiological studies showed that individual’s susceptibility to cancer is mediated by a variety of genetic factors.19 The pathogenic mechanisms of genetic factors gene polymorphisms of cytokines, growth factors, and receptors relating chronic HCV to HCC development are not entirely well established.20 VDR gene polymorphisms have been described in several chronic liver diseases such as liver cirrhosis,21 autoimmune hepatitis,22 primary sclerosing cholangitis23 and primary biliary cirrhosis.24 Moreover, it has been reported that there is a significant association between polymorphisms of VDR gene and the occurrence of HCC in alcoholic liver cirrhosis in Caucasian subjects,25 in HBV-infected patients26 and even in patients with chronic HCV.27 However, the data in the literature describing the possible association between VDR gene polymorphisms and hepatocellular carcinoma (HCC) development are scarce and inconclusive especially in patients infected with HCV. Hence, the aim was to investigate VDR gene polymorphisms as a risk for HCC development in Egyptian subjects with chronic HCV infection.
Subjects
Five hundred thirty (530) adult consecutive Egyptian outpatients attending department of internal medicine at Mansoura University Hospital with a confirmed diagnosis of hepatitis C virus infection and receiving long-term follow-up were initially enrolled in this study. Another One hundred seventy (170) age- and sex-matched healthy controls subjects were also enrolled. The age range was 18-70 years. Male to female ratio was 1.28; 393/307. The study was initiated in the October 2014 and continued through 2017. The study was approved by the Ethical Commission and Institutional review board of Mansoura Faculty of Medicine in Egypt (MFM-IRB; Code No: R/16.02.36). A written informed conscious consent was obtained from all participants before their participation. The inclusion criterion was the diagnosis of chronic hepatitis C. All patients were HCV positive. Exclusion criteria: Age below 18 years and over 70 years, a history of cancer of any type within the last 5 years, a history of solid organ transplantation or previous bone marrow transplantation, antiviral treatment and local or systemic tumor-specific treatment within the last month. Patients with chronic renal failure, bone disorders, thyroid disorders, intestinal malabsorption, previous gastrectomy, cardiac failure (ejection fraction <50%) and systemic infection (e.g. HIV, bacterial and fungal). Patients taking vitamin D, calcium or antidepressant drugs. Other causes of liver disease (e.g. alcohol consumption, chronic hepatitis B or D, primary biliary cirrhosis, metabolic liver disease, non-alcoholic steatohepatitis and autoimmune hepatitis).
Methods
Initially, all participants completed a detailed questionnaire regarding diet and habits and submitted to thorough history taking with detailed physical examinations and relevant medical history. At the day of study inclusion, three milliliters of venous blood were obtained from all participants and the serum samples were centrifuged at 3000 rpm then aliquoted and stored at -70°C until assayed. Laboratory parameters, Ultrasound, CT scans and MRI imaging model of end-stage liver disease (MELD score) and Child-Pugh scores were assessed at the time of inclusion in the study.28,29
Chronic hepatitis C was diagnosed by HCV antibody positive (third generation enzyme-linked immunosorbent assay; ELISA) and confirmed by qualitative HCV RNA polymerase chain reaction (PCR) AmplicorTM, Roch Diagnostics, Branchburg, NJ, USA. Serum HCV RNA level (Viral load) was detected using Real-Time PCR with COBAS TaqMan HCV Test (TaqMan HCV; Roche Molecular Systems Inc., Branchburg, NJ, USA; lower limit of detection: 15 IU/mL).30
Liver cirrhosis was diagnosed by ascites, esophageal varices, fundic varices, splenomegaly, jaundice, imaging and liver biopsies (if available, according to modified-knodell histological activity index).31 HCC was diagnosed by 4-phase multi-detector computed tomography (CT) scan, dynamic contrast-enhanced magnetic resonance imaging (MRI).32,33 Diagnosis of HCC was confirmed if there is one of the following three items:
25-Hydroxy vitamin D3 (25-OHD3) levels were measured using a 25-OH Vitamin D3 direct ELISA Kit intended for the quantitative determination of the 25-OHD in serum and fresh plasma (CRYSTAL CHEM, INC. Catalog. No: 09002, Assay range: 0-200 ng/mL, Assay time <2hours, Precision CV: <10%, Storage: 2-8°C).34 Samples were measured in duplicates on a Tecan SLT Rainbow plate reader (Tecan, Männedorf, Switzerland).
Detection of VDR gene polymorphisms
Genomic DNA was prepared from 2 mL EDTA-collected peripheral blood leucocytes (stored at -80oC until use) either by salting out or using Qiagen protocol (Qiagen DNA isolation kit; Wizard Genomic DNA Purification kit, Promega Corporation, Madison, WI). Genotyping was performed using a commercial PCR-SSP Sequence-Specific Primer (Olerup SSP; One Lambda Inc., Canoga Park, California, USA). VDR genotypes were identified using PCR amplification followed by restriction fragment length polymorphisms (PCR-RFLP) assay.35 Two fragments of VDR gene were amplified. The Bs fragment (825base pair) and Ap fragment (745bp). They represent two regions of VDR gene. The Bs fragment contains the BsmI restriction site with a forward primer in exon 7 and a reverse primer in intron 8. The Ap fragment contains the ApaI and TaqI restriction sites with a forward primer in intron 8 and a reverse primer in exon 9.36 The primer sequences of VDR polymorphisms at the three loci were shown in Table 1. The PCR mix contained 5μL of each primer (10pmol), 5μL buffer, 1.5μL MgCl2 (50mM), 5µL template DNA (50-100ng), 5µL dNTPs (2mmol/L), 2μL Taq polymerase (MBI Fermentas, St. Leon-Rot, Germany) and 26.5μL H2O. A total of 40 cycles of PCR were performed in a PTC-100-60 (M.J. Research, Watertown, MA, USA). An initial denaturation of DNA template at 95°C for 2 min followed by a denaturation step for 45 sec at 94°C, an annealing step for 45 sec at optimum temperature (60°C for Bs fragment and 67°C for the Ap fragment) and an extension reaction at 72°C for 1 min. After last PCR cycle, a final extension step for 2 min at 72°C was added.37 The amplified products were then digested with BsmI, ApaI and TaqI restriction endonucleases overnight at 37°C, electrophoresed on 2% horizontal agarose gel, stained with 0.5 mg/mL of ethidium bromide and visualized under UVB-illumination using the E-Gel Precast Agarose Electrophoresis System (Invitrogen Life Technologies, PA4 9RF Paisley, UK).38 The presence or absence of the desired length of PCR products determined the allelic type. The presence of BsmI, ApaI or TaqI restriction sites was defined by the lower-case ‘b’, ‘a’, or‘t’ respectively. Their absence was defined by the upper-case ‘B’, ‘A’, or ‘T’ respectively. Digestion with BsmI restriction endonuclease resulted in two fragments of 650bp and 175bp. Digestion with ApaI restriction endonuclease produced two fragments of 531bp and 214bp if the site of restriction was present. TaqI restriction resulted in three fragments of about 205, 290 and 245bp when TaqI polymorphic site is present and in two fragments of 245 and 495bp in the absence of a TaqI polymorphic site. The studied SNPs were selected on the basis of allele frequencies and functional of clinical applications.39 To improve validity and quality of genotyping, re-genotyping of 20% of samples were done by other laboratory personnel and there were no discrepancies in genotyping. Also, confirmation of genotyping of 10% randomly selected samples was done by DNA sequencing.
Single nucleotide polymorphisms (SNP) |
||
BsmI (rs1544410);
|
Forward primer |
(5’-CAACCAAGACTCAAGTACCGCGTCAGTGA-3’) |
Reverse primer |
(5’-AACCAGCGGAAGAGGTCAAGGG-3’) |
|
Exon and Intron |
7 and 8 respectively |
|
Annealing temp |
61oC |
|
Base change |
T/C (B/b) |
|
Fragments |
If restriction site present: 825, 650, 175bp |
|
If restriction site absent: 650, 175bp |
||
ApaI and TaqI; |
Forward primer |
(5’-CAGAGCATGGACAGGGAGCAA-3') |
Reverse primer |
(5’-GCAACTCCTCATGGCTGAGGTCTC-3’) |
|
ApaI ( rs7975232); |
Intron (ApaI) |
8 |
Annealing temp |
67oC |
|
Fragments |
ApaI: If restriction site present: 531, 214bp |
|
TaqI ( rs731236); |
Exon (TaqI) |
9, codon 252 |
Annealing temp. |
67oC |
|
Base change |
TaqI: G/A (T/t) |
|
Fragments |
TaqI: If restriction site present: 205, 290, 245 bp |
|
TaqI: If restriction site absent: 495, 245 |
||
Table 1 Primer Sequences, product sizes, annealing temperatures and restrictions enzymes for VDR gene genotyping
All participants were assigned to the following groups:
Statistical analysis
Data were analyzed using SPSS software (Version 17.0, SPSS Inc., Chicago, IL). Quantitative (continuous) data were expressed as (Mean ± Standard Deviation) while qualitative data and categorical variables were expressed as number and percentage. Categorical variables were compared using the chi-square (χ²) test or Fisher's exact test. Subgroups were compared using the Mc-Nemar test. One-Way Analysis Of Variance (ANOVA) followed by Post-Hoc Tukey’s honest significant difference (HSD) test was applied for multiple comparisons. ardy-Weinberg (H-W) equilibrium was used to assess the independent segregation of alleles.40 This H-W equilibrium was then assessed using chi-square (χ²) test or Fisher’s exact test when appropriate. Allele frequency was calculated as the number of occurrences of the test allele in the population divided by the total number of alleles. Carriage rate was calculated as the number of individuals carrying at least one copy of test allele divided by the total number of individuals. Variables that achieved statistical significance with the univariate analysis were included in multiple regression analysis with a forward stepwise (likelihood ratio) to evaluate the independent factors associated with risk of HCC development. Also, the odds ratios (ORs) with 95% confidence intervals (CIs) of the association between HCC and genotypic frequencies were estimated using multiple logistic regression analysis after controlling for other covariates.41 For all statistical studies, P<0.05 was considered to be statistically significant.
Primer sequences of VDR polymorphisms at three loci of VDR gene were shown in Table 1. The base change was T/C (B/b) for BsmI, C/A (A/a) for ApaI and G/A (T/t) for TaqI. The Bs fragment contains the BsmI restriction site with one primer in exon 7 and the other in intron 8. The Ap fragment contains the ApaI and TaqI restriction sites with one primer in intron 8 (ApaI) and the other in exon 9 (TaqI).
The patient basic demographic, clinical and laboratory data in all studied groups were shown in Table 2. HCC patients had a statistically significantly higher mean age than those of control patients (P=0.0064). The mean age of chronic hepatitis and liver cirrhosis patients was not significantly different from that of HCC patients. HCC patients had a statistically significantly higher male to female ratio than other groups (P=0.023). Also, HCC patients had a statistically significantly lower platelets count, lower 25-Hydroxy-Vitamin D3, higher Serum alpha-fetoprotein, higher viral load and a more derangement of liver functions (ALT, AST, and serum albumin) than other groups (P< 0.05 for all). BMI and duration of HCV infection did not significantly differ among groups (P< 0.05).
|
Control (170) |
CH (180) |
LC (180) |
HCC (170) |
CHC (530) |
ANOVA |
Post-Hoc tukey test (a,b,c,d,e,f) |
||
Group (Number) |
Control |
Group I |
Group II |
Group III) |
All patients with CHC |
F-statistics |
P-value |
Significance at |
P-value |
Age (years) |
56.3±1.21 |
58.5±1.13 |
60.15±1.02 |
62.0±1.39 |
60.18±5.4 |
4.107 |
0.0093 |
c |
0.0064 |
Male/female: (ratio) |
88/82 (1.07) |
92/88 (0.82) |
96/84 (1.14) |
117/53 (2.21) |
305/225 (1.36) |
3.155 |
0.0283 |
cefh |
0.023 |
Body mass index (BMI) (kg/m2) |
23.6±0.47 |
23.4±0.59 |
22.5±0.4 |
22.76±0.467 |
22.9±2.4 |
1.097 |
0.3542 |
- |
˃0.05 |
AST (U/L) |
40.6±1.0 |
54.95±1.52 |
95.8±3.17 |
102.1±2.087 |
83.72±23.48 |
206.81 |
1.11e-16 |
abcdegh |
<0.001 |
ALT (U/L) |
44.45±1.57 |
104.4±3.72 |
103.0±1.8 |
117.65±3.83 |
108.19±15.97 |
124.31 |
1.11e-16 |
abcefg |
<0.001 |
Platelets (104/µL) |
23.6±0.47 |
15.28±0.49 |
11.84±0.45 |
10.88±0.63 |
12.78±3.31 |
126.83 |
1.11e-16 |
abcdeg |
<0.001 |
Serum Albumin (g/dl) |
4.6±1.0 |
3.95±1.5 |
2.6±1.9 |
2.1±2.08 |
3.05±0.41 |
147.81 |
1.11e-16 |
bcdeg |
<0.001 |
Serum alpha-fetoprotein level (ng/mL) |
9.52±0.6 |
54.95±1.52 |
67.35±5.11 |
2,040.9±93.95 |
701.48±499.31 |
458.66 |
1.11e-16 |
cefgh |
<0.001 |
25-Hydroxy-Vitamin D3 (ng/ mL) |
31.88±1.82 |
27.19±1.5 |
21.7±1.06 |
21.72±1.06 |
24.2±6.4 |
11.99 |
9.40e-07 |
bcdegh |
<0.01 |
Duration of HCV infection (months) |
- |
54.95±1.52 |
56.0±2.44 |
60.8±2.77 |
58.71±3.54 |
1.832 |
0.1694 |
- |
˃0.05 |
Log HCV-RNA (IU/mL) |
- |
5.48±0.05 |
5.77±0.08 |
5.91±0.11 |
5.68±0.39 |
6.373 |
0.003 |
eh |
<0.01 |
Table 2 Baseline demographic and characteristic features of patients in all the studied groups
Data were expressed as mean±standard deviation; LC, liver cirrhosis; HCC, hepatocellular carcinoma; CHC, chronic hepatitis C;AST, aspartate transaminase; ALT, alanine transaminase; P, probability; Post-Hoc tukey test (a,b,c,d,e,f): aSignificant difference between Control and CH; bSignificant difference between Control and LC; c Significant difference between Control and HCC; d: dSignificant difference between CH and LC; eSignificant difference between CH and HCC; f Significant difference between LC and HCC; gSignificant difference between CHC and control; hSignificant difference between CHC and HCC.
The distribution of Alleles, Genotypes frequencies of VDR gene polymorphism at BsmI, ApaI, and TaqI loci in the studied groups were demonstrated in Table 3. This distribution was in accordance with the Hardy-Weinberg equilibrium. Fisher’s exact test was used to compare frequencies of HCC patients with each group. HCC patients had a statistically significantly higher frequency of ApaI CC genotype compared to control subjects (P=0.0009), patients with chronic hepatitis (P=0.0007), or liver cirrhosis (P=0.0004).
|
|
All patient (530) |
HCC(170) |
Non-HCC (LC:180, CH:180) |
Control (170) |
Fisher’s exact test |
|||
n(%) |
n(%) |
LC n(%) |
CH n (%) |
n(%) |
P1 |
P2 |
P3 |
||
BsmI |
TT (BB) |
0 (0) |
0 (0) |
0 (0) |
0 (0) |
0 (0) |
|
|
|
TC (Bb) |
53 (10) |
15(8.8) |
25(13.9) |
13 (7.2) |
20 (11.8) |
NS |
NS |
NS |
|
CC (bb) |
477 (90) |
155(91.2) |
155(86.1) |
167(92.8) |
150 (88.2) |
NS |
NS |
NS |
|
|
T/C (B/b) |
53/1007 |
15/325 |
25/335 |
13/347 |
20/320 |
NS |
NS |
NS |
ApaI |
CC (AA) |
309(59.3) |
120(70.6) |
94(52.22) |
95(52.78) |
92 (54.12) |
0.0004 |
0.0007 |
0.0009 |
|
CA (Aa) |
187 (35.3) |
40 (23.5) |
75(41.67) |
72 (40) |
66 (38.82) |
0.006 |
0.001 |
0.001 |
AA (aa) |
34 (5.6) |
10 (5.9) |
11(6.11) |
13 (7.22) |
12 (7.06) |
NS |
NS |
NS |
|
|
C/A (A/a) |
805/255 |
280/60 |
295/101 |
262/98 |
250/90 |
0.012 |
0.003 |
0.007 |
TaqI |
GG (TT) |
0 (0) |
0 (0) |
0 (0) |
0 (0) |
0 (0) |
1 |
1 |
1 |
AG (Tt) |
60(11.3) |
17 (10) |
27 (15) |
16 (8.9) |
26 (15.3) |
NS |
NS |
NS |
|
AA (tt) |
470(88.7) |
153 (90) |
153(85) |
164(91.1) |
144(84.7) |
NS |
NS |
NS |
|
G/A (T/t) |
60/1000 |
17/323 |
27/333 |
16/344 |
26/314 |
NS |
NS |
NS |
|
Table 3 Baseline demographic and characteristic features of patients in all the studied groups
Fisher’s exact test was used. P: Probability; P1, compares HCC and Liver Cirrhosis (LC); P2, Compares HCC and Chronic Hepatitis (CH); P3, Compares HCC and control; CHC, chronic hepatitis C; LC, liver cirrhosis; HCC, hepatocellular carcinoma; CH, chronic hepatitis; NS: Not significant (P ˃0.05)
Also, the distribution of Haplotypes frequencies of VDR gene polymorphism consisting of ApaI C, BsmI C, and TaqI A alleles in the studied groups were demonstrated in Table 4. HCC patients had a statistically significantly higher frequency of CCAbAt-Haplotype compared to control subjects (P=0.0099), patients with chronic hepatitis (p=0.0003), or liver cirrhosis (p=0.003). HCC patients had a statistically significantly lower frequency of ApaI CA genotype (P˂0.01) and CAA+TAG [bat+BaT]-Haplotypes (P˂0.05) than other groups. For other single nucleotide polymorphisms (SNPs) at BsmI and TapqI loci, there were no significant associations (P˃0.05 for all).
|
|
All patient (530) |
HCC(170) |
Non-HCC (LC:180, CH:180) |
Control (170) |
Fisher’s exact test |
|||
n(%) |
n(%) |
LC n(%) |
CH n(%) |
n(%) |
P1 |
P2 |
P3 |
||
Haplotypes (ApaI-BsmI-TaqI) |
CCA (bAt) |
305 (57.5) |
118(69.4) |
97(53.9) |
90(50) |
94 (55.3) |
0.003 |
0.0003 |
0.009 |
CAA (bat) |
152 (28.7) |
34 (20) |
50 (27.8) |
68 (37.8) |
50 (29.4) |
0.105 |
˂0.001 |
0.059 |
|
TAG (BaT) |
49 (9.2) |
14 (8.2) |
20 (11.1) |
15 (8.3) |
19 (11.2) |
NS |
NS |
NS |
|
CAG (baT) |
10 (1.9) |
2 (1.2) |
4 (2.1) |
4 (2.2) |
4 (2.4) |
NS |
NS |
NS |
|
TAA (Bat) |
6 (1.1) |
2 (1.2) |
3 (1.7) |
1 (0.6) |
2 (1.2) |
NS |
NS |
NS |
|
TCG (BAT) |
4 (0.8) |
0 (0) |
2 (1.1) |
2 (1.1) |
0 (0) |
NS |
NS |
NS |
|
TCA (BAt) |
2 (0.4) |
0 (0) |
2 (1.1) |
0 (0) |
1 (0.6) |
NS |
NS |
NS |
|
CCG (bAT) |
2 (0.4) |
0 (0) |
2 (1.1) |
0 (0) |
0 (0) |
NS |
NS |
NS |
|
CAA+TAG |
201 |
48 |
70 |
83 |
69 |
0.042 |
0.009 |
0.022 |
|
|
CCA/( CAA+TAG) |
305/201 |
118/48 |
97/70 |
90/83 |
94/69 |
0.042 |
˂0.001 |
0.022 |
Table 4 N=57; Epidemiological distribution of the pathological fractures, traumatic fractures, and nonunion.
The association between SNPs of VDR gene at BsmI, ApaI, and TaqI loci and disease severity in chronic HCV infection were demonstrated in Table 5. The carriage of ApaI CC genotype had a statistically significantly higher prevalence of HCC (120/309; 38.8%) compared to ApaI CA genotype (40/187; 21.4%, P˂0.05), ApaI AA genotype (10/34; 29.4%, P˂0.05) or ApaI CA plus AA genotypes (50/187; 22.6%, P=0.002) (Figure 1). Also, the carriage of CCA[bAt]-Haplotype had a statistically significantly higher prevalence of HCC (118/305; 38.7%) compared to CAA[bat] plus TAG[BaT]-Haplotypes (48/201; 23.9%, P4=0.012) (Figure 2). The carriage of CAA[bat]-Haplotype had a lower nonsignificant prevalence of HCC (22.4%). On the other hand, BsmI and TaqI polymorphisms were not significantly related to disease severity (P=0.09, P=0.125 respectively).
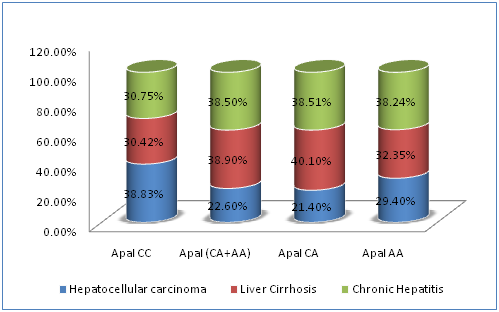
Figure 1 Correlations between different ApaI genotypes of VDR gene and disease severity in chronic HCV infection.
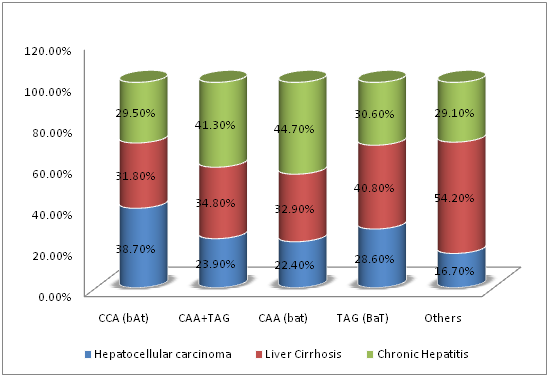
Figure 2 Correlations between different BsmI-ApaI-TaqI haplotypes of VDR gene and disease severity in chronic HCV infection.
|
HCC (170) |
LC (180) |
CH (180) |
Total CHC (530) |
p-value |
|
BsmI |
TC |
15(24.53%) |
25(47.17%) |
13(28.30%) |
53 (100%) |
P1=0.09 |
CC |
155(35.02%) |
155(32.49%) |
167(32.49%) |
477(100%) |
||
ApaI |
CC |
120(38.83%) |
94(30.42%) |
95(30.75%) |
309(100%) |
P2=0.0023 |
CA+AA |
50 (22.6%) |
86 (38.9%) |
85 (38.5%) |
221 (100%) |
||
CA |
40(21.4%) |
75(40.10%) |
72(38.51%) |
187(100%) |
|
|
AA |
10(29.4%) |
11(32.35%) |
13(38.24%) |
34(100%) |
|
|
TaqI |
AG |
17(28.33%) |
27(45%) |
16(28.33%) |
60(100%) |
P3=0.125 |
AA |
153(32.55%) |
153(32.55%) |
164(34.89%) |
470(100%) |
||
Haplotypes |
CCA (bAt) |
118 (38.7) |
97 (31.8%) |
90(29.5%) |
305 (100%) |
P4=0.012 |
CAA+TAG |
48(23.9%) |
70(34.8%) |
83(41.3%) |
201(100%) |
||
CAA (bat) |
34(22.4%) |
50(32.9%) |
68(44.7%) |
152 (100%) |
|
|
TAG (BaT) |
14(28.6%) |
20(40.8%) |
15(30.6%) |
49 (100%) |
|
|
Others |
4(16.7%) |
13(54.2%) |
7(29.1%) |
24(100%) |
|
Table 5 Correlations between SNPs of VDR gene at BsmI, ApaI, and TaqI loci and disease severity in chronic HCV infection
SNPs, single nucleotide polymorphisms; HCC, hepatocellular carcinoma; LC, liver cirrhosis; CH, chronic hepatitis; CHC, chronic hepatitis C; Chi-square was used to compare the carriage rate of genotypes in each SNPs of VDR gene. P1, Compare the Carriage Rate of BsmI TC and CC Genotypes; P2, Compare the carriage rate of ApaI CC and (CA+AA) genotypes; P3, Compare the carriage rate of TaqI AG and AA genotypes; P4, Compare the carriage rate of CCA haplotype and (CAA+TAG) haplotypes
Univariate analysis revealed that old age, male gender, lower platelets count, carriage of ApaI CC genotype and carriage of CCA[bAt]-haplotype were the factors significantly associated with HCC development. Multivariate logistic regression analysis was done after adjustment for age, sex, and platelets count to compare HCC and non-HCC patients with healthy control subjects (Table 6). There was a significant relationship between the carriage of ApaI CC genotype (using ApaI CA/AA as a reference), the carriage of ApaI C allele (using ApaI A allele as a reference) and the risk of HCC development. Also, there was a significant relationship between the carriage of CCA[bAT]-haplotype (using CAA/TAG[bAT]/BAT-haplotype as a reference) and the risk of HCC development. Comparing HCC to healthy controls, the adjusted OR (95%CI, P-value) was 2.03 (1.3- 3.2, p1=0.0018) for ApaI CC genotype, 1.7 (1.16- 2.4, P1=0.006) for ApaI C allele, 1.8 (1.14-2.85, P1=0.011) for CCA[bAT]-haplotype. Comparing non-HCC to healthy controls, the adjusted OR (95%CI, P-value was 2.17 (1.47-3.2, p2=0.0001) for ApaI CC genotype and 2.01 (1.35-2.99, p2=0.0006) for CCA[bAT]-haplotype. Comparing HCC to non-HCC patients, the adjusted OR (95% CI, P-value) was 0.46 (0.31-0.68, p3=0.0001) for ApaI CC genotype, 0.58 (0.417-0.797, P3=0.0009) for ApaI C allele and 0.497 (0.33-0.74, p3=0.0006) for CCA[bAT]-haplotype. On the other hand, there were no significant relationships between other SNPs (BsmI and TaqI) and the risk of HCC development (P˃0.05).
|
|
Control |
HCC |
Non-HCC |
HCC vs. Control |
|
Non-HCC vs. Control |
|
HCC vs. Non-HCC |
|
|
|
N (%) |
N (%) |
N (%) |
Adjusted OR (95%CI) |
Adjusted P1 value |
Adjusted OR (95%CI) |
Adjusted P2 value |
Adjusted OR (95%CI) |
Adjusted P3 value |
BsmI |
TC |
20(11.76) |
15 (8.82%) |
38 (10.56%) |
1.00* |
|
1.00* |
|
1.00* |
|
|
CC |
150(88.24) |
155(91.18) |
322 (89.44%) |
1.37(0.68-2.79) |
0.374 |
1.13(0.64-2) |
0.678 |
1.22(0.65-2.3) |
0.536 |
|
T |
20(5.9) |
15 (4.4) |
38 (5.3) |
1.00* |
|
1.00* |
|
1.00* |
|
|
C |
320(94.1) |
325(95.6) |
682(94.7) |
1.35(0.68-2.69) |
0.387 |
1.12(0.64-1.96) |
0.686 |
1.21(0.66-2.23) |
0.547 |
ApaI |
CA+AA |
78(45.9) |
50 (29.4%) |
171 (47.5%) |
1.00* |
|
1.00* |
|
1.00* |
|
|
CC |
92(54.1) |
120 (70.6) |
189 (52.5) |
2.03(1.3-3.2) |
0.0018 |
0.94(0.65-1.4) |
0.728 |
2.17(1.5-3.2) |
0.0001 |
|
A |
90(26.5) |
60(17.6) |
195(27.1) |
1.00* |
|
1.00* |
|
1.00* |
|
|
C |
250(73.5) |
280(82.4) |
525(72.9) |
1.7(1.16-2.4) |
0.006 |
0.969(0.724-1.3) |
0.834 |
1.73(1.25-2.4) |
0.0009 |
TaqI |
GA |
26 (15.3) |
17 (10) |
43 (11.9) |
1.00* |
|
1.00* |
|
1.00* |
|
|
AA |
144(84.7) |
153(90) |
317(88.1) |
1.63(0.85-3.12) |
|
1.33(0.79-2.25) |
0.286 |
1.22 (0.67-2.2) |
0.659 |
|
G |
26(7.6) |
17(5) |
43(5.97) |
1.00* |
|
1.00* |
|
1.00* |
|
|
A |
314(92.4) |
323(95) |
677(94.03) |
1.57(0.84-2.96) |
0.1659 |
1.3(0.79-2.16) |
0.303 |
1.21(0.68-2.2) |
0.523 |
Haplotype |
CAA+TAG |
69(40.6) |
48(28.2) |
153(42.5) |
1.00* |
|
1.00* |
|
1.00* |
|
|
CCA |
94(55.3) |
118(69.4) |
187(51.9) |
1.8(1.14-2.85) |
0.011 |
0.897(0.62-1.31) |
0.573 |
2.01(1.3-2.99) |
0.0006 |
Table 6 Multivariate regression analysis of factors associated with HCC risk in patients with chronic HCV infection
P1 value represents the HCC compared to healthy controls; P2 value represents the non-HCC compared to healthy controls; P3 value represents the non-HCC compared to HCC patients; P value was adjusted for age, sex, and platelets count by logistic regression analysis; HCC, hepatocellular carcinoma; CI: Confidence Interval; OR: Odds Ratio. P is significant at (P ˂0.05); * Reference allele, genotype or haplotypes
The comparisons between ApaI genotypes (ApaI CC vs. CA+AA) and [BsmI-ApaI-TaqI] Haplotypes (CCA vs. CAA+TAG) were shown in Table 7. Patients with ApaI CC genotypes and patients with CCA[bAT]-haplotype had a statistically significantly higher prevalence of HCC, pre-existing liver cirrhosis and a significantly lower serum 25-Hydroxy-Vitamin D3 concentrations (P<0.05 for all). HCV viral load did not significantly differ in patients carrying ApaI CC genotypes or CCA[bAT]-haplotype. Also, there was a higher but not significant ratio of BsmI and TaqI genotypes in patients carrying ApaI CC genotype and CCA haplotype (P >0.05).
|
ApaI genotypes |
BsmI-ApaI-TaqI haplotypes |
||||
ApaI CC |
ApaI (CA&AA) |
P-value |
CCA (bAt)-haplotype |
Other haplotypes |
P-value |
|
N |
309 |
221 |
305 |
225 |
||
Age (years) |
56.5±4.4 |
57.2±3.14 |
0.139 |
59.2±3.5 |
58.6±2.4 |
0.337 |
Male gender N (%) |
192/309 (62%) |
113/221 (51%) |
0.212 |
188/305 (62%) |
117/225 (52%) |
0.273 |
BMI (kg/m2) |
23 ±2.5 |
22.7.±2.4 |
0.059 |
22.9±2.4 |
22.8±2.5 |
0.138 |
AST (U/L) |
86±24 |
84.5±26.3 |
0.615 |
86.2±23.3 |
80.4±23.4 |
0.556 |
ALT (U/L) |
109.4±16.3 |
106.5±15.4 |
0.065 |
108.5±16.5 |
105.3±15.6 |
0.066 |
Platelets (104/µL) |
11.7±3.4 |
12.97±3.2 |
0.053 |
12.5±3.4 |
13.1±3.1 |
0.395 |
Log HCV-RNA (IU/mL) |
5.7±0.4 |
5.68±0.37 |
0.327 |
5.7±0.38 |
5.7±0.39 |
0.446 |
BsmI CC genotype |
297 (96%) |
180(81%) |
0.221 |
285(93%) |
192 (85%) |
0.482 |
TaqI AA genotype |
300 (97%) |
170 (77%) |
0.08 |
295 (93%) |
175 (93%) |
0.106 |
25-OH-VITD (ng/mL) |
21.4±5.5 |
26.3±6.1 |
0.034 |
22.0±3.5 |
25.8±6.6 |
0.047 |
HCC |
120 (39%) |
50 (23%) |
0.005 |
118 (39%) |
52 (23%) |
0.007 |
HCC with underlying LC |
120 (39%) |
28 (13%) |
˂0.0001 |
118 (39%) |
30 (13%) |
˂0.0001 |
*Total LC patients |
214 (69%) |
114 (52%) |
0.044 |
215 (70%) |
113 (50%) |
0.021 |
Table 7 Comparisons between different ApaI Genotypes and between different BsmI-ApaI-TaqI Haplotypes of VDR Gene Polymorphisms in Patients with Chronic HCV Infection (530 patients),br>Data were expressed as mean± standard deviation or number (%); LC, liver cirrhosis; HCC, hepatocellular carcinoma; AST, aspartate transaminase; ALT, alanine transaminase; AFP, serum alpha-fetoprotein level; 25-OH-VITD, 25-hydroxy-vitamin D3; BMI, body mass index; HCV, Hepatitis C Virus; P, probability; Chi-Square or Mann-Whitney U test were used. *Total LC patients (328) included 180patients with LC and 148 HCC patients with underlying LC
The carcinogenesis of HCC is a complex, multistep and multifactorial process. Chronic HCV infection, a major health problem worldwide, is ultimately complicated by HCC.20 Several genetic and epidemiological studies evidenced the pleiotropic feature of vitamin D. Recently, several VDR gene polymorphisms have been demonstrated in chronic liver diseases but with a so limited influence on the function and signaling of VDR protein.42 Also, there is scarcity in the data describing the possible association ion between VDR gene polymorphisms and HCC development. Therefore, studies are needed to clarify and understand the biological pathways, functional influences and consequences of these polymorphisms.
In this study, the possible association between VDR Gene polymorphisms and risk of HCC development in Egyptian populations infected with HCV was investigated. This study showed that patients with HCC had a significantly higher frequency of ApaI A allele, ApaI CC genotype and CCA[bAT]-Haplotype compared to control subjects (P < 0.05). For BsmI and TapqI polymorphisms, there were no significant associations (p˃0.05 for all).
Moreover, the implications of BsmI, ApaI, and TaqI polymorphisms on the severity of chronic liver diseases of HCV patients were also investigated in the current study. The carriage of either ApaI CC genotype or CCA[bAT]-Haplotype had a significantly higher prevalence of HCC, pre-existing liver cirrhosis and a significantly lower serum 25-Hydroxy-Vitamin D3 concentrations (25-OHD) compared to other genotypes or haplotypes (P <0.05 for all). On the other hand, other SNPs polymorphisms (BsmI and TaqI) were not related to disease severity (p> 0.09).
In addition to environmental, metabolic and viral factors, VDR genes genetic polymorphisms have been related to chronic liver disease and HCC development. Many authors investigated the association of several polymorphisms in VDR gene with the severity of chronic liver disease. The commonest studied genetic variations in VDR gene were the FokI, BsmI, ApaI, TaqI, and bat-haplotypes consisting of BsmI, ApaI and TaqI.43,44 There are obvious ethnic and racial variations of allelic frequencies of these VDR gene polymorphisms in different hepatic and extrahepatic diseases. BsmI, ApaI, and TaqI SNPs are located in the 3'region of VDR gene in linkage disequilibrium with each other. These polymorphisms are implicated in modulation of several chronic liver diseases such as autoimmune hepatitis (FokI polymorphisms), primary biliary cirrhosis (BsmI polymorphism), primary sclerosing Cholangitis (ApaI polymorphisms)45 and viral hepatitis.46
Also, these VDR gene polymorphisms were reported to be associated with risk HCC development in patients with liver cirrhosis,47 chronic hepatitis B,48 alcoholic cirrhotic patients underwent liver transplantation25 and chronic hepatitis C.27 Huang et al.,49 demonstrated that VDR gene polymorphisms at BsmI, ApaI, and TaqI were associated with distinct clinical phenotypes of hepatitis B carriers in Taiwanese patients but not with the risk of HCC suggesting a limited role of VDR gene polymorphisms in hepatocarcinogenesis.49 However, a biochemical evidence clearly indicated that HCC cells respond to the inhibitory effect of vitamin D and its analogs.50 Moreover, it had been reported that the antiproliferative effects of vitamin D against HCC cells correlate with intracellular VDR level.44,51 Also, it had been reported that polymorphism at FokI locus was associated with increased HCC susceptibility in Egyptian patients infected with chronic hepatitis B and had a significant role in the determination of its clinicopathological characteristics.48,26 VDR gene SNPs (ApaI CC genotypes, CCA[bAT]-haplotype and Taq AA genotype) were also reported to be significantly associated with high hepatitis C virus load but not with antiviral (peginterferon plus ribavirin) response in chronic HCV Asian patients.52 In our study, although HCC patients had significantly higher viral load than other groups, HCV viral load did not significantly differ in patients carrying ApaI CC genotypes or CCA[bAT]-haplotype.
Similar to the current study, both plasma vitamin D levels and VDR gene polymorphisms (ApaI CC genotype and bAt[CCA]-haplotype) had been reported to be significantly associated with rapid fibrosis progression rate and the presence of cirrhosis .53. Moreover, the current study further demonstrated a significant association with HCC development in CHC patients.
VDR gene polymorphism (bAt[CCA]--haplotype) was also reported to significantly impair the response to peginterferon/ribavirin-based therapy in patients with HCV and, similar to our results, could genetically predispose to low serum 25-OHD3 levels.54-56
Another study revealed that some genetic variations in binding protein (GC), CYP2R1, and DHCR7 were associated with HCC progression in HCV patients and provided an evidence for a functionally relevant contribution of reduced serum 25-OHD3 concentrations to HCV-related HCC.57
Also, it had been previously reported that reduced serum levels of 25-hydroxy vitamin D3 were linked to the HCC development; nevertheless, the causal relationships remained ambiguous because of the small sample size of these studies or the false positive associations due to impaired liver functions at the date of HCC.57 Consistent with our results, some authors reported that VDR gene polymorphisms at ApaI locus had a role in HCC development in HCV-infected patients .27. In this study, the significant association between polymorphisms in VDR gene which mediates and modulates vitamin D biological effects and HCV-related HCC could justify the causal relationships between genetic variations and distinct clinical phenotypes and suggest that an impaired vitamin D metabolism contributes to hepatocarcinogenesis in chronic hepatitis C.
In this study, the clustering in specific haplotypes (bat-haplotypes consisting of BsmI, ApaI, and TaqI), estimations of 25-OHD plasma levels and the association with a specific tumor (HCC) and/or patient characteristics might increase the clinical relevance of VDR genotypes. However, it might become more relevant when associated with SNPs of other genes or enzymes involved in vitamin D metabolism, such as binding protein (GC) and CYP enzymes (CYP27A1, CYP27B1, CYP24A1, CYP2R1).18,58
So, further controlled clinical trials are justified to validate our observations and evaluate the impact of vitamin D supplementation on HCC risk and overall survival in patients with chronic HCV. Also, the influence of ethnicity on the distribution of VDR genetic polymorphisms associated with risk of HCC should be demonstrated in further studies.
The present study provided a significant association of VDR gene polymorphisms (ApaI CC genotype and bAt.CCA-haplotype) with HCC development and disease severity in HCV-infected Egyptian patients. Also, an evidence for a functionally relevant contribution of reduced serum levels of 25-hydroxy vitamin D3 to HCV-related HCC was demonstrated in this study. Thus, the determination of VDR genetic variations in HCV patients could help identification of patients at risk of HCC development.
Authors thank Prof. Dr. Salah El-Gamal, and Dr. Aya Mohammed Amin for their support and help.
The authors declare no conflicts of interest
All the authors conceived, organized, drafted, reviewed, and approved the manuscript.
None.

©2017 Mohammed, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.