eISSN: 2379-6367


Review Article Volume 2 Issue 6
Pharmaceutical Sciences, Midwestern University College of Pharmacy, USA
Correspondence: Charles A Veltri, Midwestern University, USA, Tel 635723589, Fax 6235723565
Received: April 17, 2015 | Published: November 30, 2015
Citation: Veltri CA. Proteases: nature’s destroyers and the drugs that stop them. Pharm Pharmacol Int J. 2015;2(6):222-230. DOI: 10.15406/ppij.2015.02.00044
Proteases are found in all life forms and regulate many cellular pathways. Proteases have been proven as viable drug targets. This paper reviews the mechanism of hydrolysis of the amino acid based aspartic, cysteine, serine and threonine proteases and will also highlight the current anti protease therapeutics in the clinic.
Keywords: protease, inhibitor, drug development
Proteases are found in all life forms. Initially proteases were described from gastric juices and thought to be involved in nonspecific degradation of proteins. While some proteases are nonspecific, others are highly specific both in their substrate selection and cleavage recognition sites. Proteases account for ~2% of the human genome with 553 defined members1,2 and 1-5% of the genomes in infectious organisms such as bacteria, parasites and viruses.3 Proteases regulate many cellular processes, such as protein degradation,4,5 digestion6,7 blood coagulation,8 wound healing,9 ovulation,10 embryonic development,11 bone formation,12 neuronal outgrowth,13 cell-cycle regulation,14 immune and inflammatory response,15 angiogenesis16 and apoptosis.17 Proteases are viable drug targets because of their role in cellular functions of both mammalian cells and pathogens.
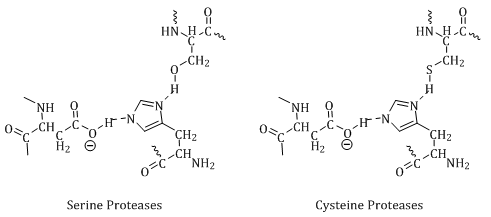
There are five major classes of proteases categorized by the catalytic method used to hydrolyze peptide bonds: serine (30%), aspartic (4%), cysteine (26%), threonine (5%) and metallo proteases (34%).3 Although the specific residues differ between the various classes, they all bind their substrates in a groove where peptide bond hydrolysis occurs, and all proteases recognize β-strands in their active site.3 The amino acids of the substrate occupy recognition pockets in the groove of the protease. The interactions are designated P3, P2, P1, P1′, P2′ and P3′ for the substrate and S3, S2, S1, S1′, S2′ and S3′ for the binding pockets. Prime and nonprime designations are with respect to the cleaved peptide bond (Figure 1). The S pockets of the protease have amino acids complementary to the side chains of the substrate at the site of cleavage. This complementation allows for selectivity in the peptide bonds cleaved by an enzyme, for example trypsin specifically cleaves at the carboxyl terminus of arginine or lysine residues.
This paper will review the mechanism of how proteases hydrolyze peptide bonds and discuss the similarities and differences between the serine, aspartic, cysteine and threonine proteases. All four of these classes of proteins share common active site functionality that is often the target for inhibitors. The development of inhibitors and their mechanism of inhibition against proteases will also be discussed. Finally, this paper will highlight the current therapeutics available on the market to target the various protease classes.
Mechanism
Serine, aspartic, threonine and cysteine proteases share common active site functionality in a nucleophilic residue coupled with a general base. The general base abstracts the proton from the nucleophilic residue to allow the active site residue to become charged (Figure 2). These residues are typically the target for inhibitors.
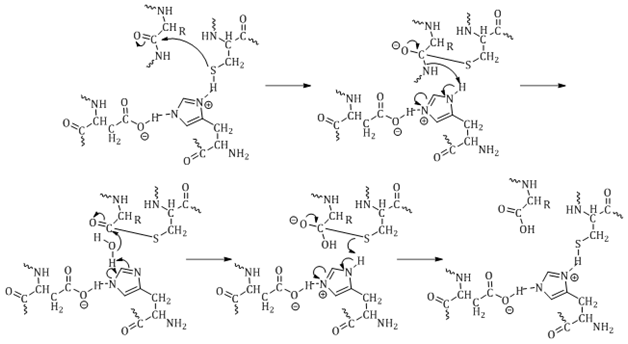
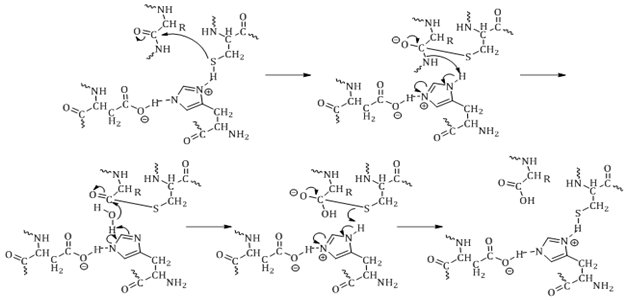
The mechanism of hydrolysis is highlighted in Figure 3, using the active site of a cysteine proteasesas the example. The protease active site residue attacks the carbonyl of the peptide bond to form a tetrahedral transition state intermediate. Electrons from the oxygen of the carbonyl drive the cleavage of the peptide bond, which abstracts hydrogen from the general base. Water then releases the second half of the product from the active site residue. The oxyanion hole generated during the transition state is stabilized through hydrogen bonding with amino groups of the backbone. The role of the oxyanion hole is not fully understood.18
While the chemistry of the mechanism of hydrolysis is similar for serine, aspartic, cysteine and threonine proteases, the amino acids involved in catalysis are what define the various classes. The different classes of proteases are discussed below.
Serine proteases
Serine proteases19‒21 use an active site serine to hydrolyze a peptide bond. The other amino acids involved in catalysis vary between the different serine protease groups. The Ser-His-Asp triad can be replaced with Ser-His-Glu, Ser-His-His or Ser-Lys depending on the family of serine protease.20,21 The serine proteases are categorized by tertiary structures into the super families of the trypsin-like or subtilisin-like.2,19,20 Examples of the trypsin-like serine proteases, the larger of the super families, are trypsin, chymotrypsin, thrombin, factor IX a, and factor Xa.2,19,20 Interestingly, some cysteine proteases adopt the trypsin-like structure.22 Examples of the subtilisin-like serine proteases are proteinase K and thermitase.2,19,20
Along with tertiary structure, serine proteases are designed against the nature of the P1 residue recognized by the protease types. The trypsin-like proteases cleave positively charged residues (Lys or Arg are preferred at P1), elastase-like proteases prefer small hydrophobic residues (Ala or Val are preferred at P1) while chymotrypsin-like proteases prefer large hydrophobic residues (Phe, Tyr, or Leu).2,19,20
Aspartic proteases
Aspartic proteases19,23‒26 use two aspartic acid residues to hydrolyze a peptide bond. These proteases are categorized into the super families of pepsin-like or viral retropepsin-like due to the tertiary structure.23,24,26,27 The pepsin-like include pepsin, cathepsin D and chymosin.23,27 The viral retropepsin-like include the retro pepsins of the immune viruses HIV, FIV and SIV.24,26
Aspartic proteases are unique in the fact they do not have a His residue in their active site. The general acid-base mechanism proposed for peptide hydrolysis involves two Asp residues and uses water to attack the targeted peptide bond (Figure 4). The peptide bond undergoes nucleophilic attack by water which is activated by the deprotonated Asp residue. The protonated Asp residue undergoes nucleophilic attack by the amide of the peptide bond, resulting in the cleavage of the peptide bond.19,23‒26
Cysteine proteases
Cysteine proteases12,18,19,22,25,28‒30 use a cysteine residue to hydrolyze a peptide bond. These proteases are categorized into three super families of viral, papain-like or caspase-like.19,22 The viral proteases have examples from many viral origins.22 As stated earlier, the viral cysteine proteases take on the serine protease trypsin-like tertiary structure.22 The papain-like proteases include the papain and cathepsins. The caspase-like proteases include the caspases and gingipain.22
Threonine proteases
The only enzymes that are threonine proteases are the proteasomes.4,5,18,31,32 Proteasomes are multifunctional and possess three distinct cleavage recognition sites: trypsin-like (Lys or Arg are preferred at P1), chymotrypsin-like which prefer large hydrophobic residues (Phe, Tyr, or Leu is preferred at P1), and caspase-like (Asp or Glu are preferred at P1).4,5,18,31 Inhibition of the proteasomes causes cell cycle arrest and apoptosis.
The proteasomes are all barrel shaped protein complexes that recognize ubiquitin-tagged proteins destined for degradation.4,5,31 Inhibiting the proteasomes can stabilize the cell cycle or lead to cell cycle arrest or apoptosis.4,5,31 It has been shown that cancer cells are more susceptible to the pro apoptotic effect of proteasome inhibitors than normal cells.5,31 This has led to the development of proteasome inhibitors as anticancer agents.
While there are many types of inhibitors for nucleophilic proteases, this section will highlight only a few alkylating agents.
Inhibitor design background
Early inhibitors of nucleophilic proteases were designed as peptide substrates to deliver a warhead to the active site.25 This approach was favorable because a specific substrate could be synthesized for the target protease. The early warheads were diazo- or halo-ketone compounds, which are excellent alkylating agents because diazo and halogens represent good leaving groups.25 The active site residue attacks the carbonyl of the warhead rather than that of the peptide bond (Figure 5).25,33
While many proteases are not highly substrate specific (only a few residues are recognized for cleavage), some proteases have extended substrate binding grooves and require longer peptides (up to 30 residues) for effective binding.25 The caspases, thrombins, neutrophil elastases and deubiquitinating enzymes are examples of proteases that recognize longer peptide fragments.4,9,25,34‒37 Therefore, the traditional method of attaching a warhead onto a substrate can become quite difficult due to the size of the peptide-warhead library necessary to identify the best inhibitors.
Two approaches have been taken to overcome this barrier. First, libraries of small molecules have been screened for inhibition of specific proteases.6,28 Screening libraries in a high-throughput assay gives hits quickly. The hits can then be further developed for greater selectivity. One important aspect of screening a library for activity against proteases is to quickly identify compounds that are nonselective alkylating, acylating, phosphorylating or sulfonating agents through confirmation assays and remove them from further study. To avoid following nonselective hits, good confirmation and secondary assays need to be employed for the specific protease of interest.
The second approach to developing protease inhibitors is to specifically design an inhibitor to the known structure of the target protease active site through computer modeling a process often referred to as in silico drug design and discovery.38,39 The amino acid sequences of 2000 proteases have been compiled into a database, MEROPS (http://merops.sanger.ac.uk/), and structured into families.19,20,22 From this database, scientists can obtain information about the secondary structure of the protease. Information from the MEROPS database coupled with structures from X-ray crystallography can be used to understand the interactions necessary for selective inhibition of proteases.38,39 This approach will increase in usage as high-throughput X-ray crystallization and structure determination are developed.40
Alkylating agents
Alkylating agents are one type of protease inhibitor class which transfers an alkyl group onto the protease active site residue to inhibit activity.18,39,41,42 Electrophilic alkylating agents add the equivalent of an alkyl cation to a nucleophile. This section will discuss three types of electrophilic alkylating agents: halomethyl ketones, diazomethyl ketones, and Michael acceptors.30
Halomethyl ketones were among the first active site-directed protease inhibitors. In the early 1960s N-tosyl-l-phenylalanine chloromethyl ketone and N-tosyl-l-lysine chloromethyl ketone (Figure 6) were developed as specific inhibitors of the serine proteases chymotrypsin and trypsin, respectively.
α-Chloromethyl ketone inhibition of serine proteases is well understood, and different from the proposed mechanism against cysteine proteases, in that α-chloromethyl ketones irreversibly bind to the His residue in the active site of serine proteases. First the active site serine attacks the carbonyl to activate the inhibitor followed by the formation of an epoxide which is then opened by nucleophilic attack on the α-carbon by His to form a tetrahedral intermediate with both the active site Ser and His residues.18 The electrons from the oxygen reform the carbonyl and release the Ser (Figure 7).
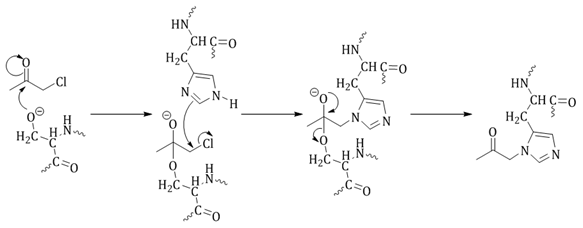
α-Chloromethyl ketones have been shown to inhibit cysteine proteases, but the exact mechanism of inhibition is not well understood.18 Inhibition of cysteine proteases results in alkylation of the active site cysteine in the form of a thioester (Figure 8). There are two potential mechanisms of inhibition: 1) direct displacement of the halogen by the thiolate anion; or 2) through a thiohemiketal followed by displacement of the halogen by the formation of a three-membered sulonium ring which rearranges to give the final thioether product (Figure 8).18
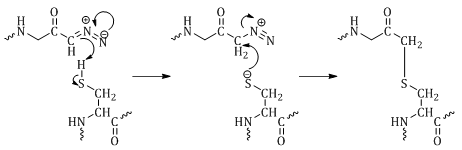
Diazomethyl ketones irreversibly bind to the active site thiol of cysteine proteases (Figure 5). Azaserine, the first compound described to inhibit cysteine proteases, was found to inhibit an enzyme involved in purine biosynthesis by alkylation of a cysteine residue.18,29 The mechanism is either through proton transfer from His to the diazo carbon adjacent to the carbonyl of the inhibitor, loss of N2 followed by alkylation of Cys, or the Cys thiol attacks the carbonyl of the inhibitor which then rearranges to the more stable thioether.18,29
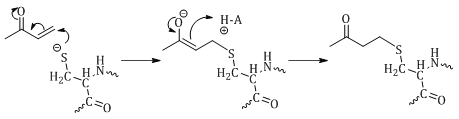
The final mechanism of inhibition discussed in this paper is that of Michael acceptors. The Michael reaction involves nucleophilic attack of and α, β-unsaturated carbonyl (Figure 9).29,42 Michael acceptors can bind either reversibly or irreversibly to the active site thiol of cysteine proteases depending on α, β-unsaturated carbonyl system.29,42 An example of a reversible Michael acceptor is α, β-unsaturated ketone.29,42
The mechanism of inhibition for Michael acceptors is well understood and involves attack on the β-carbon by the thiol of the active site Cys (Figure 8). The α-carbon is then protonated to form the thioether. Reversibility of this reaction is driven by the acidic nature of the proton on the α-carbon. If this proton is acidic, as is the case in ketones, it can be abstracted to reform α, β-unsaturated ketone resulting in breakage of the thioether bond and release of the Cys.29,42

Marine derived protease inhibitors
The idea that natural products convey some type of survival advantage is widely accepted. If natural products are synthesized to gain a survival advantage, it is reasonable to assume they have a natural target. Marine organisms are rich sources of potential drug leads because of the variety of chemistry they produce. This section will highlight marine natural products that have shown activity against proteases, specifically as anticoagulants.
Marine natural products that have anticoagulant activity interact with serine proteases such as thrombin and the factor X family. They have been found in many types of organisms from sponges,43,44 to algae,45,46 to sea cucumbers,47,48 fish49 and clams.50 It should be noted that all these marine natural products are sulfates. The sesterterpene sulfate halisulfate, isolated from Coscinoderma mathewsi, was found to be active in inhibiting thrombin and Trypsin.43 Halisulfate inhibition of thrombin and trypsin is moderate ranging from 2-100μg/mL.43
The peptides dysinosin B, dysinosin C and dysinosin D were isolated from the sponge Lamellodysidea chlorea and were shown to inhibit the serine proteases factor VIIa and thrombin.44 The three peptides have different activity against factor VIIa and thrombin. Dysinosin B is the most active against both thrombin and factor VIIa with Ki values of 0.170μM and 0.090μM, respectively. The loss of the sugar in dysinosin C results in a decrease in activity with Kis of 0.124μM and 0.550μM against factor VIIa and thrombin, respectively. The loss of the sulfate in dysinosinD drastically reduces the inhibition of thrombin from sub-μM to>5.1μM.44
The increase in activity suggests that the sugar and sulfate group both interact with the serine proteases. The large sugar may be acting as a bulky group and filling one of the S binding pockets of thrombin and factor VIIa. Since both thrombin and factor VIIa are trypsin-like serine proteases, they prefer to cleave after a positively charged amino acid at P1. The preference of a positive charge at P1 suggests the sulfate is not interacting with the active site Ser. Since sulfate groups are good hydrogen bond donors and acceptors, they may be forming hydrogen bonds with other amino acids in the active site. If the sulfates are forming hydrogen bonds with noncritical amino acids, these compounds may be nonspecific inhibitors of serine proteases.
Serine protease inhibitors in the clinic
Inhibitors of serine proteases have been designed for treatment of thrombosis, hepatitis C, angina, systemic inflammatory response, chronic obstructive pulmonary disease, pancreatitis, cancer and diabetes.9,18,34‒37,41,51‒54
Thrombin is a trypsin-like serine protease pivotal in blood coagulation and platelet aggregation. Since the 1950s, the major treatment of thrombosis has been vitamin K agonists such as warfarin. However, there are drawbacks to using warfarin, such as a narrow therapeutic range and many adverse interactions.41 These factors result in a limited efficacy of warfarin and were the driving force for the development of orally available, small molecule inhibitors of thrombin.37,55 As of 2005, two drugs have been approved for use as antithrombotic agents with eleven other compounds in various stages of preclinical and clinical trials.41
The drugs available in the clinic are described below:
The development of melagatran and ximelagatran are early examples of computer modeling to direct the synthesis of drug leads.34,36,56 An excellent review describing the 20year development of these drugs has been published.56 Melagatran is a noncovalent competitive inhibitor of thrombin. It is a potent inhibitor (Ki 2nM), but lacks selectivity over other serine proteases (KiTrypsin 3.6 nM). Melagatran is not metabolized in the body with ~80% being cleared by the kidneys without modification, and also has low oral bioavailability (~5%).34 The combination of high renal clearance with low bioavailability results in low plasma concentrations of the drug. The goal of drug therapy is to maintain plasma concentrations in the therapeutic range. Melagatran would require either large or multiple doses to maintain therapeutic levels.
Ximelagatran, a prodrug, was developed to increase the bioavailability of melagatran.56 Ximelagatran was the first orally bioavailable serine protease inhibitor to reach the clinical market.56 Adding an ethyl ester functional group to the acid increased the lipophilicity 170times over melagatran, but decreased the inhibition of thrombin (Ki 360nM). The prodrug is absorbed and metabolized to melagatran and increases the bioavailability to ~20%. The increased bioavailability increases the clinical significance of ximelagatran by maintaining therapeutic levels of melagatran for a longer period of time.
Argatroban is used for the treatment of thrombosis, vascular disease thrombocytopenia, cerebral ischemia and stroke.57 Argatroban is a potent non-covalent competitive inhibitor of thrombin (Ki 39nM) and has been used in Japan since the early 1980s in a 64:36 mixture of 21-(R) and 21-(S) diastereomers. Argatroban must be used intravenously due to its lack of oral bioavailability and short half-life (t1/2=40min).57 Argatroban is primarily cleared through the liver after metabolism by CYP3A4, or excretion through bile.58,59 The intravenous administration and short half-life do not make argatrobanan ideal drug. The ideal drug formulations are orally available for ease of patient use and have longer half-lives for fewer dosing regiments.
Two broad spectrum serine protease inhibitors have been developed for the treatment of pancreatitis. Camostat has inhibitory activity toward proteases such as thrombin, trypsin, kallikrein, plasmin, as well as other proteins such as C1 esterase, phospholipase A2 and is able to inhibit prostaglandin synthesis.51‒54 Camostat inhibits the trypsin activity in the bile and has been used in the clinic for several years in the treatment of acute pancreatitis and has been shown to control the immune response in lung injury caused by acute pancreatitis.41
Nafamostat is a naphthamidine derivative of camostat. Nafamostat has been widely used in Japan for the treatment of acute pancreatitis.41 Nafamostat is 1000X more potent than camostat against tryptase, Ki values of 95pM and 95nM, respectively. However, nafamostat has been reported to cause hyperkalemia due to reduced excretion of potassium.41 The use of broad spectrum inhibitors as therapeutic agents increases the number of diseases the drug may be used to treat, but also increases the possibility of side effects.
Aspartic protease inhibitors in the clinic
Inhibitors of aspartic proteases have been designed for treatment of AIDS, hypertension, stroke, inflammation, asthma, osteoporosis, the common cold and arthritis.26,28,41,60‒62
Inhibitors of aspartic proteases are the largest class of antiprotease drugs in the clinic. As of 2005 eight drugs were available and 27 compounds were in various stages of preclinical and clinical development.41 The large focus on inhibitors of aspartic proteases is due mostly to the disease targets of HIV/AIDS and Alzheimer’s.
Aspartic proteases are a good example of the X-ray crystallography approach to develop drug leads. Hundreds of crystal structures have been deposited in the PDB database for viral aspartic proteases (http:/www.rcsb.org/pdb/). This allows for specific inhibitors to be thoughtfully designed through computer modeling in silico prior to synthesis in wet chemistry labs.
Saquinavir was the first example of an HIV protease inhibitor obtained from structure-based design.24,26,63‒68 At the time, Phe-Pro substrates of HIV protease were not thought to be substrates for mammalian proteases, and therefore inhibitors of HIV protease were developed around this structure. Saquinavir is a penta peptide mimic with a hydroxy ethamine in place of the cleavable peptide bond; a decahydroquinoline replaces the P1′ proline; and phenylalanine is in the P1 position. Saquinavir inhibits both HIV-1 and HIV-2 replication in cell culture at low nM concentrations. Early animal experiments did not show ideal kinetics. A large dose (10mg/kg) was needed in rats, but blood levels were maintained at >15nM over more than 6 h so the drug proceeded into clinical trials. Saquinavir was approved by the FDA in 1996 under the name Invirase®, but suffered from low bioavailability (1-4%) which led to the development of a soft gel formulation, Fortovase®. Fortovase® has an 8-fold increase in bioavailability and is currently co administered with other antiretroviral drugs for the treatment of HIV.
Ritonavir is a potent inhibitor of HIV protease (Ki 15pM). Ritonavir inhibits HIV replication in the nM range (EC50 30 nM, EC90 130nM) but does not cause cell death of uninfected MT4 cells (IC50>50M).69 Ritonavir contains a hydroxyethyl transition state mimic and has terminal thiazole rings and hydrophobic Phe and Val residues which increase the bioavailability. The dosing of rats at 10 mg/kg resulted in 78% oral bioavailability; however, it binds strongly to plasma proteins (>98% of the drug is bound to plasma proteins). Ritonavir inhibits CYP3A and is often co-administered to decrease the clearance and metabolism of other HIV protease inhibitors. CYP3A is the largest subfamily of CYPs in humans. CYP3A4 metabolizes ~60% of current drugs. Co-administration of ritonavir will increase the half-life of other drugs metabolized by CYP3A through competitive inhibition of the two drugs’ metabolism.
Lopinavir, an analog of ritonavir, is 10-fold more active against HIV protease (Ki 1.3pM) and also effective against ritonavir-resistant HIV strains.70‒72 Co-administration of lopinavir with ritonavir showed a decrease of viral loads in a 3 week Phase II trial.70‒72
Indinavir is a potent inhibitor of both HIV-1 and HIV-2 proteases, Ki 0.52 and 3.3nM, respectively. Indinavir features a novel 2-aminoindanol at the P2 position and a unique transition state mimic. Press releases from Merck claim that co-administering indinavir with ritonavir and two non-nucleoside reverse transcriptase inhibitors reduces viral load in patients treated for either 48 or 100weeks.68,69,73
Nelfinavir was developed to couple the bulky P1′ substituent and hydroxyethylamine transition state mimic of saquinavir with an extended P1 substituent to produce a more lipophilic inhibitor. Phase II trials suggest that nelfinavir was the best tolerated first generation HIV protease inhibitor and displayed acute viral reductions of >90% and was approved for use by the FDA in 1997.68,69,74,75
Amprenavir uses the bulky P2 substituent idea of saquinavir, but contains a sulfonamide in place of the hydroxyethylamine. Amprenavir is a water soluble, potent inhibitor of both HIV-1 protease (Ki 600pM) and HIV replication (EC90 80nM). During a Phase III clinical trial, amprenavir dosed at 1.2g twice a day for 3weeks coadministered with two non-nucleoside reverse transcriptase inhibitors reduced viral loads compared to patients not given amprenavir.60,62,75‒77 Amprenavir is given in large doses, but has a long half-life (7-10h)60,62,75‒77 which means the patients need fewer doses. Amprenavir was approved for use by the FDA in 1999.75‒77
HIV is a difficult disease to develop drugs for. The HIV proteases mutate rapidly and develop resistance to drugs (some within days to weeks).41 The need for more effective anti-HIV drugs drives the development of second generation HIV protease inhibitors. The ideal second generation anti-HIV drug will have increased potency toward the HIV protease, high oral bioavailability, longer half-life, and increased plasma concentration.
Fosamprenavir is a phosphate derivative of amprenavir. Fosamprenavir has similar potency against HIV-1 protease and bioequivalence to amprenavir, but is more water soluble and has a higher oral bioavailability.78 Fosamprenavir does meet the criteria of a better second generation drug in the fact that it is orally available, but since it does not exhibit an increase in activity, fosamprenavir will most likely face similar resistance problems as the first generation anti-HIV drugs.
The second generation HIV protease inhibitor, atazanavir is a tetrapeptide mimic with a 2-hydroxy-1,3-diaminopropane transition state mimic, an aza-dipeptide core and an extended P1′. Atazanavir inhibits HIV-1 protease (Ki 10pM) and HIV infected peripheral blood mononuclear cells (IC50 8-12nM), and inhibits indinavir-resistant and saquinavir-resistant HIV-1 strains, IC50 3-100nM and 4-100nM, respectively.75,78‒80 A dose of 300mg of atazanavir resulted in plasma levels greater than the EC50 value for over 24h.75,78‒80 The >24h time period of effective drug levels in the plasma permits single daily dosing. Single daily dosing is unique for anti-HIV medications which often require multiple daily doses.
Cysteine protease inhibitors in the clinic
Cysteine protease inhibitors have been designed for the treatment of osteoarthritis, inflammation, hepatic dysfunction, myocardial infarction, cerebral ischaemia, severe acute respiratory syndrome and the common cold.12,18,25,29,30,41,42
Inhibitors have been designed against the preferred recognition site of these types of enzymes, but there are no anticysteine protease drugs available in the clinic.41 The lack of clinically available cysteine protease inhibitors has left an open niche in drug discovery. However, studies have been performed to better understand the mechanism of action of the punaglandins, marine natural products, which have been shown to inhibit cysteine proteases.81,82
Threonine protease inhibitors in the clinic
The only threonine protease inhibitor in the clinic is bortezomib, marketed by Millennium Pharmaceuticals, Inc. as Velcade™.41 Three other drug leads are in various stages of development.41
Bortezomib is an anticancer agent used in the treatment of relapsed and refractory multiple myeloma 131,41,83 Bortezomib is a potent and selective inhibitor of the chymotrypsin-like activity of the proteasome, with a Ki 620 pM toward the 20S proteasome.41,83 Bortezomib is a reversible competitive inhibitor that binds to the threonine hydroxyl of the active site through the boronic acid (Figure 10).31
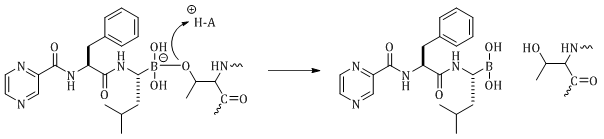
The boron atom covalently binds to the oxygen of the active site threonine to form a borate. The reaction is reversible to relieve the negative charge of the borate (Figure 10). The fact that the boron binds to the oxygen suggests that bortezomib will also inhibit serine proteases, but bortezomib is selective for the proteasomes.41,83 Bortezomib forms additional hydrogen bonds in the chymotrypsin-like and caspase-like active threonine proteases sites of the 20S proteasome when compared to serine proteases.
The proteasome develops resistance over time and the compound does have side effects in humans, including diarrhea, neurotoxicity, fatigue, fever, anorexia, nausea, vomiting, rash and headache (administered at a dose range of 0.13-1.56mg/m2/dose).83 The effectiveness of bortezomib in the treatment of cancer has proven the proteasome pathway as a viable drug target. The toxicity and development of resistance with bortezomib suggests that discovering inhibitors of other proteins in the pathway may lead to treatments for multiple myeloma with fewer side effects.
Proteases are essential enzymes for many biological processes. Proteases utilize the same chemistry of nucleophilic and basic residues to hydrolyze peptide bonds. The difference in the active site residues of serine, aspartic, cysteine and threonine proteases affords scientists with different targets to design specific inhibitors. For example, the serine, cysteine and threonine proteases all use a His residue as the basic amino acid in their active sites, but aspartic proteases use a deprotonated Asp as a conjugate base. Specific inhibitors can be designed against the protease classes by targeting the different amino acids in the active site of the various protease families.
Many approaches have been exploited in the design of protease inhibitors. Peptide mimics of substrates have been used to deliver warheads to the active site of proteases. This approach to the design of inhibitors has been used historically, but can be time consuming and costly. Two approaches have been utilized to decrease the development time of new protease inhibitors: a screen of small molecule libraries and computer modeling to guide the design of potential inhibitors before their synthesis. In the future, technology could help develop focused screens for later generation inhibitors. One example would be to couple molecular modeling and X-ray crystallography with selective natural product libraries to increase the development of future generations of protease inhibitors.
Despite a significant effort, very few drugs are clinically available in contrast to the large number of proteases. Clinically available serine protease inhibitors are used to treat thrombosis and pancreatitis even though serine proteases regulate more biological functions than blood clotting and digestion. Inhibitors designed against aspartic proteases dominate the clinical market with eight drugs available as of 2005.41 The large number of drugs that target aspartic proteases is due to their use as anti-HIV medications. However, HIV proteases mutate rapidly and thereby develop resistance to the medications quickly. There are no clinically available drugs against cysteine proteases, and only one clinically available drug that targets threonine proteases. The small number of clinically available protease inhibitors gives potential for drug development.
I would like to express my deepest appreciation to Tim S. Bugni and Mary Kay Harper for their diligent reading and critiques of this paper.
Author declares that there is no conflict of interest.

©2015 Veltri. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.