eISSN: 2379-6367


Research Article Volume 11 Issue 1
1Research Center in Biodiversity and Genetic Resources (CIBIO/InBIO), University of Porto, Portugal
2School of Medicine and Biomedical Sciences of the University of Porto (ICBAS-UP), Portugal
3I3S-Instituto de Investigação e Inovação em Saúde, Universidade do Porto, Portugal
4Ipatimup-Institute of Molecular Pathology and Immunology of the University of Porto, University of Porto, Portugal
5Department of Pathology, Faculty of Medicine, University of Porto, Portugal
6Faculty of Medicine, University of Coimbra, Portugal
7Coimbra Hospital and University Center (CHUC), Portugal
Correspondence: Eliane Silva, Research Center in Biodiversity and Genetic Resources (CIBIO/InBIO), University of Porto, Vairão, Portugal
Received: January 01, 2023 | Published: January 9, 2023
Citation: Silva E, Marques S, Osório H, et al. Occult hepatitis C infection: viruses with infectious potential in Huh7.5 and MDBK cell lines suggest HCV/OCI transmission. Pharm Pharmacol Int J. 2023;11(1):1-8. DOI: 10.15406/ppij.2023.11.00394
Background: Occult hepatitis C infection (OCI) is characterized by the detection of HCV-RNA in patient’s peripheral blood mononuclear cells (PBMCs) without detection in serum. We aim to evaluate the capacity of viruses presented in OCI patients to infect and replicate in Huh7.5 (human hepatoma–derived Huh7 cell line) and in naïve Mardin-Darby Bovine Kidney (MDBK) cell lines questioning HCV/OCI transmission.
Methods: Huh7.5 cells were infected with serum and PBMCs samples of 4 OCI patients. Naïve MDBK cells were infected with the same serum samples. Huh7.5 and naïve MDBK cells were also infected and reinfected, with an inoculum of naïve MDBK cell cultures previously infected with the same 4 serum samples, respectively. All infected/reinfected Huh7.5 and MDBK cell cultures were screened for HCV/OCI-RNA detection by droplet digital-PCR (ddPCR). MDBK cell cultures infected with the serum samples were evaluated for HCV/OCI specific proteins by proteomic liquid chromatography-mass spectrometry (LC-MS) analysis. Huh7.5/infected and MDBK/reinfected cell cultures with the inoculum serum samples were also evaluated for the E2, NS3 and NS4a HCV/OCI specific regions by immunogold electron microscopy (IEM) analysis.
Results: HCV/OCI-RNA was detected in Huh7.5 cell cultures infected with serum and PBMCs samples of the OCI patients 4 and 3, respectively, and in Huh7.5/infected and MDBK/reinfected cell cultures with the inoculum serum samples of the 4 OCI patients by ddPCR. HCV specific proteins peptides were detected in MDBK cell cultures (first infection) infected with 1 serum sample by LC-MS. HCV specific immunostaining was shown in Huh7.5 cell cultures infected with PBMCs samples of the OCI patients’ 3- NS3 and 4-NS4a, and in MDBK cell cultures reinfected with inoculum serum sample of the OCI patient 4-E2 and -NS4a by IEM analysis.
Conclusions: Infectious potential of HCV/OCI viruses was demonstrated in Huh7.5 cell cultures infected with serum and PBMCs samples of the 4 OCI patients. Infectious potential of HCV/OCI viruses was also demonstrated in MDBK cell cultures infected with the serum samples, as well as, in Huh7.5/infected and MDBK/reinfected cell cultures with the inoculum serum samples. Overall results suggest HCV/OCI transmission, and could contribute for a better understanding of the HCV/OCI life cycle, infection and transmission.
Keywords: HCV, OCI, Huh7.5 cell line, MDBK cell line, HCV/OCI transmission, Droplet digital PCR, LC-MS, IEM
Occult hepatitis C infection (OCI) is characterized by the detection of hepatitis C virus (HCV)-RNA in liver biopsy patient’s samples and in peripheral blood mononuclear cells (PBMCs) without detection in serum samples.1-3 It is a common condition worldwide, all HCV genotypes can be involved in this form of infection,4 and no HCV vaccine is currently available.5 OCI has been described in patients with chronic liver disease, coinfections, comorbidities and in population with risk factors, such as, in patients infected with hepatitis B virus or human immunodeficiency virus, with tattoos, drugs users, and apparently in healthy population.6-11 The primary goal of HCV therapy is to cure the infection, i.e. to achieve a sustained virological response (SVR) defined as undetectable HCV-RNA in serum after treatment completion, and the use of direct-acting antiviral agents (DAAs) markedly improved the SVR rates.12-14 Although, the detection of OCI in patients who achieved a SVR with DAAs was previously described,15-17 and we have recently presented OCI detection in people with or without DAAs therapy.18
HCV present tropism to replicate in the liver-hepatocytes, and also have capacity to replicate in PBMCs and lymphoid cells.19,20 While, in 2003 Kato et al cloned a HCV-2a consensus genome with the capacity to produce infectious HCV in human hepatoma-derived Huh7 cell line.21 Furthermore, Huh7.5 (human hepatoma–derived Huh7 cell line) cell line has also been widely used for the propagation of HCV in vitro, and both cell cultures demonstrated to be suitable experimental substitutes for primary hepatocytes.22-25 Moreover, the replication of HCV homologs in non-hepatoma cells, such as in mosquito, African green monkey, Vero cells, and Mardin-Darby Bovine Kidney (MDBK) cell lines have also been previously described.26-28 The aim of this study was to evaluate the capacity of viruses detected in OCI patients to infect and replicate in Huh7.5 and in naïve MDBK cell lines by droplet-digital PCR (ddPCR), proteomic liquid chromatography-mass spectrometry (LC-MS) and immunogold electron microscopy (IEM) analysis questioning HCV/OCI transmission.
Patients
OCI patients that were referred to the Coimbra Hospital and University Center (CHUC) were identified in a previous study we have performed.18 Here, we randomly select 4 OCI patients from the previously identified, patient 1, 2, 3 and 4, and performed the cell lines investigation attending to the HCV/OCI transmission possibility. Two HCV positive patients (patient 5 and 6) were also included in the study.
Samples
Blood samples from 4 OCI patients were collected in dry and in lithium heparin tubes,and serum was recovered from the dry tubes as previously described.29 PBMCs were isolated using lymphoprepTM (Alere Technologies AS, Norway) following the manufactures instructions, and recovered PBMCs were resuspended in 200 µL of water molecular grade (G-Biosciences, USA) and also in 200 µL of RPMI 1640 culture medium (Thermo Fisher Scientific, USA) supplemented with 50% fetal bovine serum (FBS) (Thermo Fisher Scientific, USA) and 20% dimethyl sulfoxide (Thermo Fisher Scientific, USA). All samples were stored at -80ºC. Then, Huh7.5 cell line was infected with serum and PBMCs resuspended in water molecular grade samples. Naïve MDBK cell line was infected with serum samples, and Huh7.5 and naïve MDBK were also infected and reinfected with an in vitro serum samples, respectively. All infected/reinfected Huh7.5 and MDBK cell cultures were screened for HCV/OCI-RNA detection by droplet digital-PCR (ddPCR). MDBK cell cultures infected with the serum samples (first infection) were evaluated for HCV/OCI specific proteins by proteomic liquid chromatography-mass spectrometry (LC-MS) analysis. Huh7.5/infected and MDBK/reinfected cell cultures with the inoculum serum samples were also evaluated for the E2, NS3 and NS4a HCV/OCI specific regions by immunogold electron microscopy (IEM) analysis. Blood samples from the 2 HCV positive patients were also collected and evaluated as for the 4 OCI patients.
Cell lines
Huh 7.5 cell line was kindly provided by Professor Charles Rice (Apath, L.L.C and Rockefeller University, USA) and maintained in Dulbecco`s Modified Eagle Medium (Thermo Fisher Scientific, USA) supplemented with 10% FBS (Thermo Fisher Scientific, USA), 0.1% penicillin (100 U/mL)–streptomycin (100 µg/mL) (GRISP, Porto, Portugal), 200 mM L-glutamine (Thermo Fisher Scientific, USA) and 1% of non-essential amino acids (Thermo Fisher Scientific, USA) at 37°C and 5% CO2, similarly as previously described.25,30 MDBK cell line was purchased at the European Collection of Authenticated Cell Cultures and were maintained as previously described.28,31
Viral infection in Huh7.5 and in MDBK cell lines
Huh7.5 cells were infected with serum and PBMCs resuspended in water molecular grade samples (4 OCI patients and 2 HCV positive patients) to evaluate the infectivity and efficient production of HCV/OCI particles following previously described methods for naïve MDBK cells infection.28,31 Naïve MDBK cell line was infected with serum samples (4 OCI patients and 2 HCV positive patients), and Huh7.5 and naïve MDBK cells were also infected and reinfected with an inoculum serum samples (4 OCI patients and 2 HCV positive patients), respectively, for the same evaluation similarly as previously described.28,31 Briefly, 2 mL of each serum and PBMCs resuspended in water molecular grade samples filtered through a 0.2 µm membrane were adsorbed onto Huh7.5 cells in T25 flasks and incubated at 37°C and 5% CO2 for 3 h. Monolayers were washed twice with 1X PBS, the culture medium was replaced, and flasks were maintained at 37°C and 5% CO2 for 7 days (S-Huh7.5-1I-P0 and PBMCs-Huh7.5-1I- P0 passages, 7 days’ post infection). After, 1 (P1) and 2 (P2) subcultures, respectively, were performed and maintained at the same conditions and for the same time, S-Huh7.5-1I-P1, PBMCs-Huh7.5-1I-P1 and PBMCs-Huh7.5-1I-P2, respectively. Supernatants and cells recovered separately from each S-Huh7.5-1I-P1 and PBMCs-Huh7.5-1I-P2 flask/sample were then evaluated for HCV/OCI-RNA detection by ddPCR, and the cell cultures from each PBMCs-Huh7.5-1I-P2 flask/sample were also evaluated for HCV/OCI specific immunostaining by IEM. Uninoculated Huh7.5 cells (negative control) were maintained and also evaluated by ddPCR and IEM.
In naïve MDBK cells a first inoculation (1I) of serum samples was performed. Two mL of each serum sample filtered through a 0.2 µm membrane were adsorbed onto naïve MDBK cells in T25 flasks and incubated at 37°C and 5% CO2 for 3 h. Monolayers were washed twice with 1X PBS, the culture medium was replaced, and flasks were maintained at 37°C and 5% CO2 for 7 days (S-MDBK-1I-P0, 7 days’ post infection). After, 1 subculture (P1) of these cell cultures was performed and maintained at the same conditions and for the same time, S-MDBK-1I-P1 passage. Supernatants from each S-MDBK-1I-P1 cell cultures flask/sample, and from uninoculated MDBK cells (negative controls) flasks were recovered and evaluated for HCV/OCI specific proteins by LC- MS analysis, and remaining cell cultures were stored at –80°C for 8 days. After, cell cultures from S-MDBK-1I-P1 were evaluated for HCV/OCI-RNA detection by ddPCR and used later as inoculum to reinfect naïve MDBK and infect Huh7.5 cell lines. For the reinfection in naïve MDBK cells and the infection in Huh7.5 cells with the serum inoculum samples, 2 mL of the inoculum from each cell culture sample were also filtered through a 0.2 µm membrane, adsorbed onto naïve MDBK and Huh7.5 cells in T25 flasks and incubated at 37°C and 5% CO2 for 3 h. Monolayers were washed twice with 1X PBS, the culture medium was replaced and flasks were maintained at 37°C and 5% CO2 for 7 days, S-MDBK-RI-P0 and S-Huh7.5-I-P0, respectively. Then 2 and 1 subcultures for the MDBK-RI-P0 and S-Huh7.5-I-P0, were performed, S-MDBK-RI-P1, S-MDBK-RI-P2 and S-Huh7.5-I-P1, respectively, and supernatants and cells recovered separately from each S-MDBK-RI-P0, S-MDBK-RI-P1, S-MDBK-RI-P2, S-Huh7.5-I-P0 and S-Huh7.5-I-P1 flask/sample were screened for HCV/OCI-RNAdetection by ddPCR. Cell cultures from each S-MDBK-RI-P2 and S-Huh7.5-I-P1 flask/sample were also evaluated for HCV/OCI specific immunostaining by IEM. Uninoculated MDBK and Huh7.5 cells (negative controls) were maintained and also evaluated by ddPCR and IEM.
A flowchart of the study design for the Huh7.5 and naïve MDBK cells infection/reinfection assays is presented in Figure 1.
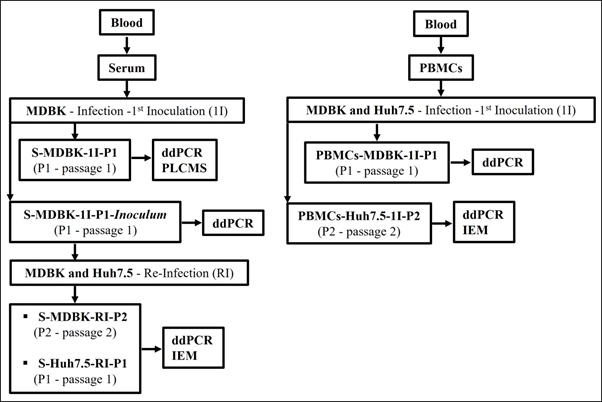
Figure 1 Flowchart of the study design for the MDBK and Huh7.5 cells infection and reinfection assays.
RNA extraction, cDNA synthesis and ddPCR
Total RNA from cell cultures was extracted using the RNeasy Mini Kit (Qiagen, Hilden, Germany) following the manufactures instructions. The cDNA synthesis was performed using random hexamers included in the Xpert cDNA synthesis kit (GRISP Porto, Portugal) following the manufactures instructions. DdPCR was performed as previously described8,32 with some modifications. Briefly, HCV/OCI-RNA was detected for the HCV core region using only the sense 5-GCGTTAGTAYGAGTGTYG-3´ and antisense 5-CRATTCCGGTGTACTCAC-3´ primers, that were able to amplify HCV genotypes 1, 2, 3 and 4.30 The reaction mix contained 10 μL of ddPCR™ Supermix for Probes (Bio-Rad, USA), 900 nM of each primer, 250 nM of FAM-labeled HCV probe (5′-FAM-CCGCAGACCACTATGGCTC-BHQ1-3′) and 2 μL of template in a final volume of 22 μL. Amplicons are of 89 bp. The reactions mix were placed on a Bio-Rad QX200 Droplet Digital PCR System for droplets generation following the manufactures instructions, and then HCV/OCI-RNA amplification was performed on a Bio-Rad Thermal Cycler C1000 (Bio-Rad, USA) under the cycling conditions of 95°C for 10 min, 45 cycles at 94°C for 30 s and 55°C for 1 min, and 98°C for 10 min, with a hold step at 4°C. After, the plate was read on a Bio-Rad QX200™ Droplet Reader for droplets analysis, and data was analysed using the Bio-Rad QuantaSoftTM Analysis Pro Software v. 1.0.596, following manufactures instructions. The fluorescence amplitude threshold was automatically adjusted in individual wells and HCV/OCI-RNA concentration was achieved in copies/μL attending to the number of observed accepted droplets (10.000 or greater). For each tested sample, two runs were performed and in duplicate to adjust ddPCR sensitivity. The plasmid pGNN-non-replicative (p7-nsGluc2A) kindly provided by Professor Charles Rice (Apath, L.L.C and Rockefeller University, USA) was used as positive control in ddPCR reactions.
LC-MS analysis
Supernatants from S-MDBK-1I-P1 cell cultures/flasks and from un-inoculated MDBK cells (negative controls) flasks were recovered and evaluated for HCV/OCI specific proteins by LC-MS analysis as previously described33 with some modifications. Briefly, 300 µL of each cell culture supernatant were recovered to 1.5 mL low protein binding microcentrifuge tubes (Thermo Fisher Scientific, USA) and were placed in a water bath at 95°C for 10 min. Protein extractions from each cell culture supernatant were performed and recovered proteins were digested with trypsin. The resulting peptides were then analysed by LC-MS. In data analysis the HCV/OCI protein peptides identification was performed with the data available in the UniProt protein sequence database for the Homo sapiens Reviewed Proteome 2021_03 with 20,371 entries, hepatitis C virus Proteome 2021_04 with 220,291 entries, Bos taurus Reviewed Proteome 2021_03 with 6,014 entries and a common contaminant database from MaxQuant (version 1.6.2.6, Max Planck Institute of Biochemistry, Munich, Germany). The mass spectrometry proteomics data have been deposited to the Proteome Xchange Consortium via the PRIDE (http://www.ebi.ac.uk/pride/archive/) partner repository with dataset identifier PXD033680 and 10.6019/PXD033680. This procedure was performed at the i3S Proteomics Scientific Platform.
IEM analysis
Huh7.5 cell cultures infected with PBMCs samples, passage PBMCs-Huh7.5-1I-P2, were evaluated for HCV/OCI specific immunostaining for the E2, NS3 and NS4a HCV specific regions by IEM analysis as previously described28,31 with some modifications. Moreover, Huh7.5 and MDBK cell cultures that were infected and re-infected with the inoculum serum samples, S-Huh7.5-I-P1 and S-MDBK-RI-P2, respectively, were also evaluated. Briefly, cell culture suspensions were fixed in 2,5% glutaraldehyde (Electron Microscopy sciences, USA) and 2% paraformaldehyde (Merck, Germany) in tris buffered saline. Cell cultures were evaluated using primary mouse monoclonal antibodies anti-E2, anti-NS3 and anti-NS4a HCV proteins at dilutions 1:10 and/or 1:25. Huh7.5 and MDBK uninoculated cells (negative controls) were evaluated using anti-E2 mouse monoclonal antibodies at dilution 1:10.
DdPCR Huh7.5 and MDBK cell cultures
Huh7.5 cells were infected with serum and PBMCs re-suspended in water molecular grade samples of the 4 OCI patients to evaluate the infectivity and efficient production of HCV/OCI. Huh7.5 cells were also infected with samples of the 2 HCV positive patients and evaluated. From these cell cultures, supernatants and cells recovered separately from each S-Huh7.5-1I-P1 and PBMCs-Huh7.5-1I-P2 flask/samples were evaluated for HCV/OCI-RNA detection by ddPCR. Considering supernatants plus cells ddPCR results, positives results were detected in S-Huh7.5-1I-P1 and in PBMCs-Huh7.5-1I-P2 infected with the 4 OCI patients’ serum and PBMCs samples, respectively, with the exception for PBMCs-Huh7.5-1I-P2-OCI patient 4 (Table 1). Positive results were also detected in S-Huh7.5-1I-P1 and in PBMCs-Huh7.5-1I-P2 infected with the 2 HCV positive patients’ serum and PBMCs samples, with the exception for S-Huh7.5-1I-P1-HCV positive patient 6 (Table 1). In S-Huh7.5-1I-P1infected with the 4 OCI patients’ serum samples, HCV/OCI-RNA was detected in a range of 0.14 to 1.89 copies/µL with 17963 and 18088 number of accepted droplets, respectively, and for the S-Huh7.5-1I-P1 infected with the sample of the HCV positive patient was of 0.14 copies/µL with 17123 number of accepted droplets (Table 1). In PBMCs-Huh7.5-1I-P2 infected with the 4 OCI patients’ PBMCs samples HCV/OCI-RNA was detected in a range of 0.21 to 0.56 copies/µL with 16953 and 16919 number of accepted droplets, respectively, and for PBMCs-Huh7.5-1I-P2 infected with the samples of the HCV positive patients was of 0.32 to 1.01 copies/µL with 15156 and 15998 number of accepted droplets, respectively, (Table 1). Negative results were obtained for the uninoculated Huh7.5 cells (negative control).
Cell cultures |
Patients |
OCI Patients |
HCV positive patients |
||||
1 |
2 |
3 |
4 |
5 |
6 |
||
SN-S-Huh7.5-1I-P1 |
HCV/OCI-RNA (Copies/μL) |
0 |
0.46 |
1.89 |
0.67 |
0.14 |
0 |
Accepted droplets |
14383 |
17963 |
18088 |
15847 |
17123 |
12251 |
|
Cells-S-Huh7.5-1I-P1 |
HCV/OCI-RNA (Copies/μL) |
0.62 |
1 |
1.08 |
0.9 |
0.14 |
0 |
Accepted droplets |
17104 |
11717 |
16288 |
16991 |
17123 |
12251 |
|
SN + Cells - S-Huh7.5-1I-P1 |
+ |
+ |
+ |
+ |
+ |
- |
|
SN-PBMCs-Huh7.5-1I-P2 |
HCV/OCI-RNA (Copies/μL) |
0.5 |
0.56 |
0.35 |
0 |
0.32 |
1.01 |
Accepted droplets |
16380 |
16919 |
13638 |
15325 |
15156 |
15998 |
|
Cells-PBMCs-Huh7.5-1I-P2 |
HCV/OCI-RNA (Copies/μL) |
0 |
0.21 |
0.39 |
0 |
0 |
0 |
Accepted droplets |
16712 |
16953 |
17780 |
12273 |
16394 |
15049 |
|
SN + Cells - PBMCs-Huh7.5-1I-P2 |
+ |
+ |
+ |
- |
+ |
+ |
|
Table 1 HCV/OCI-RNA detected in Huh7.5 cell line infected with serum and PBMCs samples of the 4 OCI and 2 HCV positive patients by ddPCR
SN, supernatant; S, serum; P, passage; PBMCs, peripheral blood mononuclear cell; HCV, hepatitis C virus; OCI, occult hepatitis C infection; (+) positive result; (-) negative result.
The S-MDBK-1I-P1 cells cultures (SN + cells) that were used as the inoculum serum to reinfect new naïve MDBK and to infect Huh7.5 cells were evaluated for HCV/OCI-RNA detection by ddPCR. HCV/OCI-RNA was detected only in the cell cultures infected with the serum of the OCI patient 3, and negative results were shown in cell cultures infected with the serum samples of the HCV positive patients (Table 2). In S-MDBK-1I-P1 cells cultures infected with the serum of the OCI patient 1 HCV/OCI-RNA was detected with 0.98 copies/µL and 15594 number of accepted droplets (Table 2). Negative results were obtained for the uninoculated MDBK cells (negative control).
Moreover, S-MDBK-RI-P0, S-MDBK-RI-P1 and S-MDBK-RI-P2 reinfected cell cultures (SN and cells recovered separately) were analysed for HCV/OCI-RNA detection by ddPCR. Considering supernatants plus cells and the 3 evaluated passages (S-MDBK-RI-P0, -P1 and -P2) ddPCR results, positive results were detected in all tested reinfected MDBK cell cultures that were infected with the MDBK inoculum serum samples-4 OCI patients (Table 2). Negative or not determined results were shown in the tested MDBK cell cultures reinfected with the MDBK inoculum serum samples-2 HCV positive patients (Table 2). HCV/OCI-RNA (S-MDBK-RI-P0, -P1 and -P2) was detected in a range of 0.15 to 3.14 copies/µL with 15771 and 15887 number of accepted droplets, respectively (Table 2). Negative results were obtained for the uninoculated MDBK cells (negative control).
Furthermore, S-Huh7.5-I-P0 and S-Huh7.5-I-P1 infected cell cultures (SN and cells recovered separately) were analysed for HCV/OCI-RNA detection by ddPCR. Considering supernatants plus cells and the 2 evaluated passages (S-Huh7.5-I-P0 and -P1), positive results were detected in all tested Huh7.5 infected with the MDBK inoculum serum samples of the 4 OCI and the 2 HCV positive patients (Table 2). HCV/OCI-RNA (S-Huh7.5-I-P0 and -P1, -4 OCI patients) was detected in a range of 0.13 to 3.44 copies/µL with 18402 and 14044 number of accepted droplets, respectively (Table 2). HCV/OCI-RNA (S-Huh7.5-I-P0 and -P1, -HCV positive patients) was detected in a range of 0.79 to 1.77 copies/µL with 16394 and 15998 number of accepted droplets, respectively (Table 2). Negative results were obtained for the uninoculated Huh7.5 cells (negative control).
Positive result was obtained for the tested plasmid pGNN-non-replicative-p7-nsGluc2A (ddPCR positive control) by ddPCR, with 22.4 copies/µl and 17423 number of accepted droplets.
Cell cultures |
Patients |
OCI Patients |
HCV positive patients |
||||
1 |
2 |
3 |
4 |
5 |
6 |
||
Inoculum serum-S-MDBK-1I-P1 |
HCV/OCI-RNA (Copies/μL) |
0 |
0 |
0.98 |
0 |
0 |
0 |
Accepted droplets |
12518 |
14868 |
15594 |
15537 |
15568 |
14942 |
|
SN-S-MDBK-RI-P0 |
HCV/OCI-RNA (Copies/μL) |
0 |
0.15 |
0 |
0 |
N.D |
N.D |
Accepted droplets |
14075 |
15771 |
15057 |
16797 |
N.D |
N.D |
|
Cells-S-MDBK-RI-P0 |
HCV/OCI-RNA (Copies/μL) |
0.28 |
0.9 |
0 |
0 |
N.D |
0 |
Accepted droplets |
16950 |
14378 |
16632 |
14375 |
N.D |
N.D |
|
SN + Cells - S-MDBK-RI-P0 |
+ |
+ |
- |
- |
N.D |
-/N.D |
|
SN-S-MDBK-RI-P1 |
HCV/OCI-RNA (Copies/μL) |
1.03 |
0 |
0 |
0 |
N.D |
0 |
Accepted droplets |
12563 |
10925 |
15799 |
12896 |
N.D |
15264 |
|
Cells-S-MDBK-RI-P1 |
HCV/OCI-RNA (Copies/μL) |
0.22 |
0 |
0.21 |
0 |
N.D |
0 |
Accepted droplets |
16265 |
14440 |
11313 |
15537 |
N.D |
16778 |
|
SN + Cells - S-MDBK-RI-P1 |
+ |
- |
+ |
- |
N.D |
- |
|
SN-S-MDBK-RI-P2 |
HCV/OCI-RNA (Copies/μL) |
3.41 |
0 |
0.49 |
2.11 |
N.D |
N.D |
Accepted droplets |
15887 |
14192 |
19027 |
17722 |
N.D |
N.D |
|
Cells-S-MDBK-RI-P2 |
HCV/OCI-RNA (Copies/μL) |
2.71 |
0 |
0 |
0 |
N.D |
N.D |
Accepted droplets |
16525 |
16893 |
17542 |
16099 |
N.D |
N.D |
|
SN + Cells - S-MDBK-RI-P2 |
+ |
- |
+ |
+ |
N.D |
N.D |
|
SN-S-Huh7.5-I-P0 |
HCV/OCI-RNA (Copies/μL) |
0 |
3.44 |
3.18 |
0.47 |
0 |
0 |
Accepted droplets |
17006 |
14044 |
15978 |
10021 |
15156 |
15998 |
|
Cells-S-Huh7.5-I-P0 |
HCV/OCI-RNA (Copies/μL) |
0 |
1.07 |
0 |
0.18 |
0.79 |
0 |
Accepted droplets |
15214 |
16737 |
12137 |
18513 |
16394 |
15049 |
|
SN + Cells - S-Huh7.5-I-P0 |
- |
+ |
+ |
+ |
+ |
- |
|
SN-S-Huh7.5-I-P1 |
HCV/OCI-RNA (Copies/μL) |
0 |
0 |
0.13 |
0 |
0 |
1.77 |
Accepted droplets |
16406 |
18102 |
18402 |
15617 |
15156 |
15998 |
|
Cells-S-Huh7.5-I-P1 |
Copies/μL |
1.54 |
0 |
0 |
0.72 |
0.79 |
0 |
Accepted droplets |
16024 |
16431 |
17466 |
16268 |
16394 |
15049 |
|
SN + Cells - S-Huh7.5-I-P1 |
+ |
- |
+ |
+ |
+ |
+ |
|
Table 2 HCV/OCI-RNA detected in infected/reinfected MDBK and infected Huh7.5 cell cultures with the serum samples of the 4 OCI and 2 HCV positive patients by ddPCR
SN, supernatant; S, serum; P, passage; PBMCs, peripheral blood mononuclear cell; HCV, hepatitis C virus; OCI, occult hepatitis C infection; (+) positive result; (-) negative result.
LC-MS in MDBK cell cultures
Supernatants recovered from the S-MDBK-1I-P1 cell cultures (first inoculation) that were infected with the serum samples of the 4 OCI and 2 HCV positive patients were evaluated by LC-MS. HCV/OCI specific protein peptide (Protein F) was detected only in the SN of the cell cultures that were infected with the OCI patient 3 serum sample (Table 3). Relatively to the HCV positive patients, HCV/OCI specific proteins peptides (Core, E1 and RdRp) were detected only in the SN of the cell cultures that were infectedwith the HCV positive patient 6 serum sample (Table 3). The detected HCV/OCI specific proteins peptides were not shown in the uninoculated MDBK cells (negative controls).
Patients |
UniProt accession no. (Genotype) |
HCV Protein |
Best peptide sequence |
Genome position (aa) |
Exp. Q-value |
3 |
A0A650C4D1 (N/A) |
Protein F |
ADPGRSLGMLGLFMAMR |
60-76 |
0 |
6 |
K0A6T5 (1b) |
Core |
FAQGWGPITYAEPPK |
465-479 |
0.005 |
6 |
A0A1B1LUY2 (N/A) |
Core |
DAIILLTCAVYPELIFDITK |
871-890 |
0.005 |
6 |
X2BYG4 (N/A) |
Core |
LGNEILLGPADTEDTAGWR |
1006-1024 |
0.006 |
6 |
A0A139ZG90 (N/A) |
Core |
LPTAAMGAAYGFQYSPKQR |
2608-2626 |
0.005 |
6 |
B1NE60 (N/A) |
E1 |
VAIIMVMFSGVDATTHTVGSSTAR |
53-76 |
0.006 |
6 |
A0A1I9RGK9 (N/A) |
E1 |
THIDMVVMSASLCSALY |
260-276 |
0.005 |
6 |
I1UZB4 (1) |
RdRp |
LQDCTMLVCGDDLVVICESAGVHEDAASLR |
90-119 |
0.006 |
6 |
A0A2H4YRE0 (N/A) |
RdRp |
LPGAVMGAAYGFQYSPKER |
182-200 |
0.005 |
6 |
B1PBT9 (N/A) |
RdRp |
LQVLDDHYQDVFK |
532-544 |
0.005 |
Table 3 HCV/OCI protein peptides identified in the supernatants of MDBK cell cultures infected with serum samples of the 4 OCI and 2 HCV positive patients by LC-MS analysis
HCV, hepatitis C virus; aa, amino acids; N/A, not available.
IEM in Huh7.5 and MDBK cell cultures
Cell cultures suspensions S-Huh7.5-1I-P1, PBMCs-Huh7.5-1I-P2 and S-MDBK-RI-P2 that were infected or reinfected with the 4 OCI and 2 HCV positive patients’ samples were also evaluated by IEM using mouse monoclonal antibodies anti-HCV E2, anti-HCV NS3 and anti-HCV NS4a. HCV/OCI specific immunostaining was shown in PBMCs-Huh7.5-1I-P2 cell cultures infected with the PBMCs samples of the OCI patients 3 (NS3) and 4 (NS4a), and serum samples of the HCV positive patient 6 (E2) (Figure 2A). In S-MDBK-RI-P2 cell cultures HCV/OCI specific immunostaining was shown in cell cultures infected with serum sample of the OCI patient 4 (E2 and NS4a) (Figure 2B). Uninoculated Huh7.5 and MDBK cells (negative controls) were also evaluated with the anti-HCV E2 monoclonal antibodies and no HCV/OCI specific immunostaining was shown (Figure 2A and 2B).
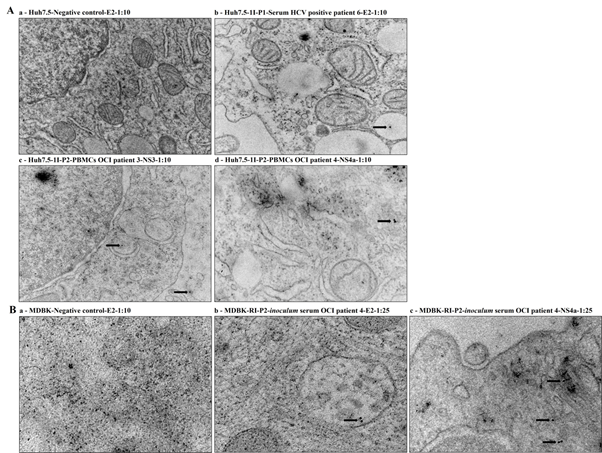
Figure 2 HCV specific immunostaining for the HCV E2, NS3 and NS4a specific regions by IEM. (A) IEM detection in Huh7.5 cell cultures infected with serum and PBMCs samples of the 4 OCI and 2 HCV positive patients. (a) Huh7.5-Negative control-E2-1:10, (b) Huh7.5-1I-P1-Serum HCV positive patient 6-E2-1:10, (c) Huh7.5-1I-P2- PBMCs OCI patient 3-NS3-1:10 and (d) Huh7.5-1I-P2-PBMCs OCI patient 4-NS4a-1:10. The immunogold particles were 10 nm in diameter. Scale bars: (a and c 0,5 µm); (b 0,2 µm); (d 200 nm). (B) IEM detection in MDBK cell cultures reinfected with inoculum serum samples. (a) MDBK-Negative control-E2-1:10, (b) MDBK-RI-P2-inoculum serum OCI patient 4-E2-1:25, (c) MDBK-RI-P2-inoculum serum OCI patient 4-NS4a-1:25. The immunogold particles were 10 nm in diameter. Scale bars: (a 200nm); (b and C 100 nm).
OCI is normally characterized by the detection of HCV in PBMCs patient samples without detection in serum samples,1-3 that could be negative due to the presence of low viral load not easily detected by the current methods and/or technologies. Furthermore, data where the possibility to detect HCV in serum samples after ultracentrifugation preventing low HCV viral load detection in OCI patients was also previously described.34,35 In this line, we have performed a previous study in OCI where we have characterized OCI patients with HCV/OCI negative and positive results in serum and PBMCs samples, respectively, by ddPCR.18 Here, we have randomly selected 4 OCI patients from our previous study and performed the work. Huh7.5 and naïve MDBK cell lines were infected with the serum of the 4 OCI identified patients in order to evaluate if serum samples could have HCV/OCI viruses that were not detected by ddPCR in our previous study. We also infect Huh7.5 cells with PBMCs samples of the 4 OCI patients that tested positive for HCV/OCI by ddPCR in our previous study. We aimed to investigate the possibility of the viruses presented in both samples to be infectious in both cell lines and understand if they could also have the capacity of HCV/OCI transmission. When Huh7.5 cell line was infected with the serum and PBMCs samples of the 4 OCI patients’ positive results were shown in all infected cell cultures by ddPCR. These results demonstrated that HCV/OCI viruses are presented in serum samples previously determined as HCV/OCI negative using the same methods and ddPCR reaction.18 Moreover, these results demonstrated also the infectious potential of the presented viruses in the serum and PBMCs samples in Huh7.5 cell line. This achievement in infected Huh7.5 cells was not previously described, while the HCV capacity to infect and replicate in Huh7.5 cell line was previously described.21-25
Furthermore, when naïve MDBK cell line was infected with the serum inoculum S-MDBK-1I-P1 cell cultures samples (first infection) of the 4 OCI patients, positive results were just detected in cell cultures infected with the serum inoculum sample of the OCI patient 3 by ddPCR. Independently of this only detected positive result, infectious potential of the presented HCV/OCI viruses were demonstrated in the infected MDBK cell line, and HCV-homologs with the capacity to infect and replicate in this cell line were previously described.28,31 Moreover, when naïve MDBK and Huh7.5 cell lines were reinfected and infected with the 4 OCI patients serum inoculum S-MDBK-1I-P1 cell cultures samples, positive results were detected in all tested reinfected MDBK and infected Huh7.5 cell cultures, meaning that serum samples of the 4 OCI patients have viruses with the capacity to infect MDBK cells, and that the viruses presented in the 4 OCI patients serum inoculum S-MDBK-1I-P1 cell cultures have also the capacity to internalize and replicate in reinfected MDBK cell cultures and in infect Huh7.5 cell cultures. These results are accordantly with the detected positive results shown in the analysed Huh7.5 cell cultures infected with the serum samples of the 4 OCI patients. Overall results demonstrated the infectious potential of the HCV/OCI viruses presented in the serum and PBMCs samples in Huh7.5 and MDBK cell lines suggesting the possibility of HCV/OCI transmission. To the best of our knowledge, no work was published describing the possibility of HCV/OCI transmission using Huh7.5 and MDBK cells infected with serum and PBMCs samples of OCI patients. Otherwise, an HCV cell-cell transmission assay using Huh7.5.1 cells was previously described.36 Moreover, nowadays the HCV life cycle remains partly understood due to the difficulty of culturing HCV in vitro or obtaining fully susceptible yet immunocompetent in vivo models, nonetheless, an HCV life cycle in cells, infection and host response, and analysis of molecular markers influencing the outcome of infection and response to therapy was described in 2016.37 Taking in consideration our previously reported data and obtained results here we suggest that our work could support in the understanding of the HCV/OCI life cycle, HCV/OCI infection and HCV/OCI transmission. LC-MS analysis were performed in supernatants recovered from the S-MDBK-1I-P1 cell cultures (first inoculation) that were infected with the serum samples of the 4 OCI patients and an HCV specific protein peptide was identified in cell cultures infected only with the serum of the OCI patient 3. This result is accordantly with the obtained by ddPCR, while more positive results were achieved using the methodology and the ddPCR reaction. Although, a proteomic analysis by LC-MS reporting the first large-scale proteome analysis of the highly permissive Huh7.5 cells containing a full-length HCV replicon was previously described,38 as well as, HCV specific proteins peptides identified in MDBK cells inoculated with rabbit and hare samples by proteomic analysis.28,31
Furthermore, HCV specific immunostaining for the NS3 and NS4a proteins was demonstrated in Huh7.5 cell cultures infected with the PBMCs samples of the OCI patient 3 and 4. In MDBK cell cultures reinfected with the serum inoculum sample-OCI patient 4 was demonstrated for the E2 and NS4a proteins. No HCV/OCI specific immunostaining was observed for all tested infected/Huh7.5 and reinfected/MDBK cell cultures, although achieved results are accordantly with the encountered by ddPCR-OCI patients 3 and 4 and LC-MS-OCI patient 3 analysis and are accordantly also with previously described data.22-25,28,31
In this study infectious potential of HCV/OCI viruses was demonstrated in Huh7.5 cell cultures infected with serum and PBMCs samples of the 4 OCI patients. Infectious potential of HCV/OCI viruses was also demonstrated in MDBK cell cultures infected with the serum samples of the 4 OCI patients’, as well as, in Huh7.5 and in MDBK cell cultures infected and reinfected with the inoculum serum samples-4 OCI patients, respectively. Overall results suggest HCV/OCI transmission and suggest also that our work could support in the understanding of the HCV/OCI life cycle, HCV/OCI infection and HCV/OCI transmission.
Informed consent was obtained from all subjects involved in the study under the project approval by the CHUC ethical committee (registration number CHUC-122-18).
None.
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (http://www.ebi.ac.uk/pride/archive/) partner repository with the dataset identifier PXD033680 and 10.6019/PXD033680.
This research was funded by the Foundation for Science and Technology (FCT) under the project PTDC/SAU-SER/30788/2017, FEDER.
Eliane Silva and Armando Carvalho designed the study. Armando Carvalho, Adélia Simão, João Madaleno and Bernardo Canhão participated in the patient’s recruitment, data collection, and data interpretation and all together with Eliane Silva participated in the sample’s recruitment. Eliane Silva performed the ddPCR and the in vitro cells works, and prepared samples for LC-MS and IEM analysis, with Eliane Silva IEM results responsibility. Hugo Osório performed the LC-MS analysis and analysed data with Eliane Silva. Eliane Silva wrote the paper with Sara Marques support. All authors critically reviewed the manuscript and approved the final version of the manuscript for publication.
The authors acknowledge the Foundation for Science and Technology (FCT) under the project PTDC/SAU-SER/30788/2017, FEDER. The authors acknowledge the Cancer Biology & Epigenetics Group, Research Center of IPO Porto, and the Molecular Pathology and Immunogenetic Laboratories, ICBAS-UP, for their availability to carry out the work. The authors also acknowledge the project NORTE-01-0246-FEDER- 000063, supported by Norte Portugal Regional Operational Programme (NORTE2020), under the PORTUGAL 2020 Partnership Agreement, through the European Regional Development Fund (ERDF) that co-funded the work.
The authors declare that they have no competing interest.

©2023 Silva, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.