eISSN: 2379-6367


Mini Review Volume 8 Issue 6
1Centre for Forensic Programmes and DNA Studies, University of Benin, Nigeria
2Department of Chemical Pathology, School of Medicine, University of Benin, Nigeria
3Edo State Command, Police State Headquarters, Nigeria
Correspondence: Nwawuba Stanley Udogadi, Centre for Forensic Programmes and DNA Studies, University of Benin, Benin City, Nigeria, Tel +234-8065699068
Received: November 29, 2020 | Published: December 15, 2020
Citation: Nwawuba SU, Momoh SM, Nwokolo CC, et al. Key DNA profiling markers for identification: A mini review. Pharm Pharmacol Int J. 2020;8(6):337-343. DOI: 10.15406/ppij.2020.08.00315
DNA Profiling also referred to as DNA fingerprinting, is a sequence of tests and methodology employed to examine and identify the genetic information contained within an individual’s cell, and within the context of forensics, it is considered as the comparison of genetic information or profile of a person to a profile of biological evidence found at the scene of crime, or profile stored in the DNA database for the sole purpose of administration of justice. Remarkably, a core base for DNA profiling is the fact that, about 99.9% sequence of DNA is found to be common among humans, and the other 0.1% is unique and vary from person to person, and the likelihood of two individual having exactly the same DNA profile is about 1 in 594.1 trillion individuals. Therefore, the present study elucidated key molecular markers for DNA profiling including short tandem repeat, variable number tandem repeat, and restriction fragment length polymorphism. In conducting this study, a comprehensive electronic literature search using PubMed, ScienceDirect, Google Scholar, and Google search for similar and related works were used, and all works meeting the subject matter were considered, including; reviews, meta-analyses, retrospective studies, observational studies, organization recommendations, and original articles. Going forward, the stated molecular markers have been demonstrated to be used for identification purpose, however, the validated technology of short tandem repeat and also its abundance in the human genome makes it a better option for forensic DN profiling and it is currently the gold standard for analysis and storage of DNA profiles in the DNA databases around the world.
Keywords: DNA profiling, forensic DNA profiling, short tandem repeat, variable number tandem repeats, restriction fragment length polymorphism
DNA Profiling also referred to as DNA fingerprinting, is a sequence of tests and methodology employed to examine and identify the genetic information contained within an individual’s cell.1 In the context of Forensics, Forensic DNA, a fortuitous byproduct of some of the most highly regarded, cutting-edge research in the scientific community, is regarded as the comparison of genetic information or profile of a person to a profile of biological evidence found at the scene of crime, or profile stored in the DNA database for the sole purpose of administration of justice.1,2 Since the use of DNA fingerprinting technique in 1986 by Dr. Alec J. Jeffreys for the identification and conviction of pitchfork for the rape and murder of 15 years old Dawn Ashworth, it has revolutionized the criminal justice system and courts around the world now considers DNA evidence acceptable and admissible.3
The basis of DNA profiling/fingerprint is on the fact that every person has a unique set of genetic fingerprints that trails us, and this unique genetic fingerprint is a hereditary blueprint transmitted from parents to offspring.4 The genetic information is stored in a molecule called the deoxyribonucleic acid (DNA), as a code made up of four nucleotide bases including: Adenine (A), Guanine (G), Cytosine (C), and Thymine (T) of which, adenine and guanine are regarded as double-ringed purines, while cytosine and thymine are regarded as single-ringed pyrimidines.2 The target of most of the molecular markers in use for DNA profiling is reading the nucleotide sequences within the DNA of a person’s cell. Therefore, the present study discusses molecular markers currently in use for identification purpose which includes; Restriction Fragment Length Polymorphisms (RFLP), Variable Number of Tandem Repeat Sequences (VNTR) Typing, and particularly, Short Tandem Repeat (STR) typing which is the gold standard for generation of profiles for storage in the DNA database.
Going forward, Short tandem repeats (STRs), also referred to as microsatellites, are short fragments of tandemly repeated DNA sequences of two to six base pairs in length found at a specific location in the chromosome mostly at center, called centromere.4-6 At the specific chromosomal location, a locus is found to contain two allelic short tandem sequences of DNA transferred by each parent of an individual; Variable number tandem repeat (VNTRs) is also known as the minisatellite consisting of core repeated sequences of greater than six (> 6) mostly sequences of about (20-100 bases) repeated between 0 to 30 times. Minisatellites repeats are found in the genome organized as tandem repeats flanked by segments of non-repetitive sequences.7 They are found to be located on many chromosomes, and the number of tandem sequences repeats in a certain locus are vastly variable among unrelated persons;8 and Restriction fragment length polymorphism (RFLP) is a molecular marker that utilizes the enzyme restriction endonuclease which cuts DNA strands at a specific site called the restriction site to produce fragments of DNA or restriction fragments of defined length.9 Finally, the stated molecular markers have been demonstrated to be used for identification purpose, however, the validated technology of short tandem repeat and also its abundance in the human genome makes it a better option for forensic DNA profiling and it is currently the gold standard for analysis and storage of DNA profiles in the DNA databases around the world.
This review was carried out by a comprehensive electronic literature search using PubMed, ScienceDirect, Google Scholar and google search. The following key words and their combination: “Forensic DNA Profiling”, “Molecular Markers”, “Short Tandem repeat”, “Restriction Fragment Length Polymorphism”, “Variable Number Tandem Repeat” and “Deoxyribonucleotide (DNA)”. All works meeting the subject matter were considered, including; reviews, meta-analyses, organization recommendations, and original articles. Preference was placed on current papers nonetheless did not exclude frequently referenced and highly regarded older publications.
An overview of Deoxyribonucleic Acid (DNA)
Long before the discovery of DNA which stands for deoxyribonucleic acid, some body of scientific evidence revealed that DNA was too small a molecule to carry out life’s biological instructions. Rather, scientist believed that the greater complexity and structural form of the polymer protein were likely to carry out vital biological functions.3,10 Following the debate on the molecule with genetic/hereditary material and vital biological function, several experimentations were conducted and novel scientific findings emerged. Summarily, a crystal understanding of the key role of the DNA as the genetic material was evident after the novel scientific breakthrough of Alfred Hershey and Martha Chase in 1952.3,10 Additionally, in 1953, the works of the following scientist; James Watson, Francis Crick, Maurice Wilkins and Rosalind Franklin, on X-ray diffraction patterns, elucidated the double helix structure of DNA; a structure that allows it to transmit biological information from one generation to the next.10
The information in DNA is stored as a code made up of four nucleotide bases including: Adenine (A), Guanine (G), Cytosine (C), and Thymine (T) of which, adenine and guanine are regarded as double-ringed purines, while cytosine and thymine are regarded as single-ringed pyrimidines.2 This nucleotide bases pair up obeying the Chargaff’s role, A bonding to T by a double bond and G bonding to C by a triple bond, to form the units termed base pairs.3 Structurally, each base is attached to a ribose sugar molecule, and a phosphate group; which confers a negative charge on the DNA, to form a nucleotide. Nucleotides are arranged in two long stands running anti-parallel, one from 3’ to 5’ end and the other from 5’ to 3’ end to form a spiral called a double helix. In general, the double helix structure of the DNA is to a certain degree like a ladder, with the nucleotide pairs forming the ladder’s rungs and the ribose sugar and the phosphate group forming the vertical sidepieces of the ladderas demonstrated in figure 1.10

Figure 1 The Structure of the DNA.10
DNA is the genetic material in humans and virtually all other organism. The human genome contains about three (3) billion bases with well over 99 percent of the bases similar in all people. It is found in almost all cells of the human body exception of the red blood cells, with its predominance located in the cell nucleus (nuclear DNA) but a small fragment can as well be found in the mitochondria called the (mitochondrial DNA or mtDNA).11 An important property of the DNA is the ability to replicate, and the replicative ability is critical during cell division, and also enables the genetic instruction contained in the DNA to be transmitted from parents to offspring’s during reproduction.10 Except for homozygous twins, the genetic instructions contained in the DNA is unique to each person and it is an omnipresent residue that trails us wherever we go.3
Based on the hierarchy of structural complexity, it follows thus; cell, nucleus, chromosome and then the DNA as demonstrated in figure 2. In this light, the DNA, a threadlike structure is found in the nucleus of the human cell, tightly wound extensively around the histone proteins named chromosomes.4,12 Archetypally, the chromosomes consist of two segments called arms which joins at a constriction point called the centromere. The two segments are regarded as the long and short arms of the chromosome represented by ‘q’ and ‘p’ arms respectively. Note worthily, the location of the centromere is found very useful in describing the location of a specific gene.4

Figure 2 A typical cell structure, nucleus, chromosome, and DNA.14
The human cell contains a pair of sex chromosomes and 22 pairs of autosomes, and based on distinctiveness, the prominent variation between the autosome and the sex chromosome is that, the former controls the sex and related individual traits, but the later regulates the somatic characteristics of a person.4,13 Going forward, the two-sex chromosome in humans are called allosomes, and it is chiefly responsible for the determination of an individual’s sex. In this setting, the two humans sex chromosomes are represented as ‘X’ and ‘Y’ with the females containing two homomorphic copies of ‘X’ chromosome arranged in the same order, while males, carries a heteromorphic copies of sex chromosomes namely ‘X’ and ‘Y’ chromosomes.4,12
Forensic DNA profiling
DNA Profiling also referred to as DNA fingerprinting, is a sequence of tests and methodology employed to examine and identify the genetic information contained within an individual’s cell.1 In the context of Forensics, Forensic DNA, a fortuitous byproduct of some of the most highly regarded, cutting-edge research in the scientific community, is regarded as the comparison of genetic information or profile of a person to a profile of biological evidence found at the scene of crime, or profile stored in the DNA database for the sole purpose of administration of justice.1,2 One of the core bases for DNA profiling is the fact that, about 99.9% sequence of DNA are found to be common among humans, and the other 0.1% is unique and vary from person to person.3 The likelihood of two individual having exactly the same DNA profile is about 1 in 594.1 trillion individuals,15 and as result, forensic DNA profiling has outstandingly contributed in exonerating the innocent and convicting the guilty.16
In addition, an estimated 30,000 genes are found to spread in approximately 3 billion base pair of the DNA in the human genome, and these genes are encoded in approximately 5% of the human genome.3 The greater part of the human genome is predominated by the “non-coding region” with well over 90% of which do not carry the genetic information necessary for the gene expression, and as a result, it is found to be very suitable for forensic DNA profiling because they are informative for individual variability and identification.17 A large body of scientific findings have demonstrated that genetic variation is rather limited in the coding region with an exception to the human leukocyte antigen (HLA) region.17 This is as a result of the fact that expressed genes are subject to a selection pressure through the course of evolution in order to maintain their specialized function. Whereas, the non-coding region is not mainly influenced by selection pressure, and as such, there is a generational transmission of mutation leading to a great increase in genetic variability.17,18
A significant part of the non-coding region of about 50% contains repetitive sequences broadly divided into two classes including; tandem repeat sequence and intersperse elements. In the forensic settings, most of the DNA fingerprinting techniques targets the tandem repetitive sequences.17 Forensic DNA profiling approach has been corroborated to be reliable, reproducible, accurate, and founded on validated methods for equally generating DNA profile and the interpretation of that profile,3,8,19 and as such, forensic DNA evidence has been granted a transcendent and epistemic status in many courts around the world.3,20,21
Since the conviction of Pitchfork using evidence from forensic DNA profiling methodology, the admissibility and acceptance of DNA evidence around the world have continued to grow, and it is considered as a gold standard in criminal investigation.22,23 Historically, in 1986, Dr. Alec J. Jeffreys was requested by the UK police to conduct a DNA profiling of a suspect, Richard Buckland who had admitted and made confessional statement about the rape and murder of 15 years old Dawn Ashworth.3,24 In profiling the samples from a crime scene in 1983 and 1986, the genetic professor, Dr. Alec J. Jeffrey found out that, although the samples matched each other, but samples profile was not consistent with the Richard Buckland’s DNA profile.3,8,24 In an effort to discover the actual culprit, the UK police commenced a genetic dragnet; collecting samples from well over 4,000 men between the age of 17 and 34 in Leicestershire, yet a consistent profile was not found.3 Colin Pitchfork who had evaded the genetic dragnet was eventually arrested, profiled, and his DNA profile matched the crime scene sample. On the 19th September, 1987, he was convicted and sentenced to life imprisonment, and was the first murderer to be convicted based on irrefutable evidence from DNA profile.24
Following the pitchfork case in 1987, substantial resources has ever since then been committed in developing and refining the DNA profiling methodology and technology.3 Presently, DNA profiling technology and methodology in estimating the frequencies have developed to a stage where the admissibility of appropriately collected and profiled DNA data should not be in doubt.3,8,25 Going forward, a prelude to the method for DNA profiling would be instructive. DNA profiling or fingerprinting in forensics commonly includes the following steps.15
DNA profiling markers
Methodologies and technologies in use for Forensic DNA profiling varies in their differentiating ability between two persons, the rapidity and sensitivity of the obtained result. In recent times, it has been demonstrated that there is a huge improvement in the speed of performing forensic DNA profiling, as DNA profiling that previously took well over one week is now conducted in a few hours.17 In this light, of course polymorphic proteins biomarkers and enzymes previously used, were found to be infrequent and provided only limited identification information.26 This led to innovations which sprouted the sophisticated tools for DNA profiling. Since the discovery of a reliable and modern methodology for the generation of a genetic profiles, and its ability for pinpointed individual identification in the criminal investigation settings by Sir Alec Jeffreys, developments in DNA profiling methodology and technology have allowed for a speedy and specific generation of tremendously discriminating profiles.27,28 Therefore, the molecular technology/markers currently in use for DNA profiling include the following but not necessary all; Restriction Fragment Length Polymorphisms (RFLP) Method of DNA Profiling, Variable Number of Tandem Repeat Sequences (VNTR) Typing, and particularly, Short Tandem Repeat (STR) typing which is the gold standard for generation of profiles for storage in the DNA database.
Short tandem repeat
Short tandem repeats (STRs), also referred to as microsatellites, are short fragments of tandemly repeated DNA sequences of two to six base pairs in length found at a specific location in the chromosome mostly at centre, called centromere.4-6 At the specific chromosomal location, a locus is found to contain two allelic short tandem sequences of DNA transferred by each parent of the individual. Going forward, these short fragments of nucleotide bases are found be constantly repetitive, and the number of repeats has been demonstrated to be individualized.23 Short tandem repeats are widely distributed in the human genome, accounting for about 3% of the entire human genome with its predominance in the noncoding region of the genome. Within this distribution, STRs are mostly found in the noncoding regions, but can also slightly be found in the coding.5,6
Based on varying repetitive sequences, microsatellites are categorized as follows; according to the length of major repeated sequence which are classified into; mono, di, tri, tetra, penta, and hexanucleotide repeats, and also according to the repetitive structure; classified into perfect repeats, regarded as simple repeats and mostly contains a single repetitive unit, and imperfect repeats regarded as compound repeats and it is characterized by more than one repetitive units.5,29 The wide distribution of microsatellite and the uniqueness with location specificity, necessitated a uniform and simple nomenclature for the human identification. For instance, the STR marker designated as “D3S1266”, D represents DNA, 3: Chromosome 3 on which the STR locus locates, S: STR, and 1266: theunique identifier.3,5
Due to the highly polymorphic nature of STRs, the higher the number of STRs loci used for the purpose of identification, the greater the discriminating power. 3,23 In addition, the likelihood of two person from a random population pool possessing the exact number of repetitive units, remains extremely rare except for homozygotic twins.8 For instance, at the same locus, a tetra-nucleotide repeat sequence (represented by CTAG) varies between various person as shown in figure 3. Person one (1) has 5 repeats, person two (2) has 6, and 7 repeats in three (3).
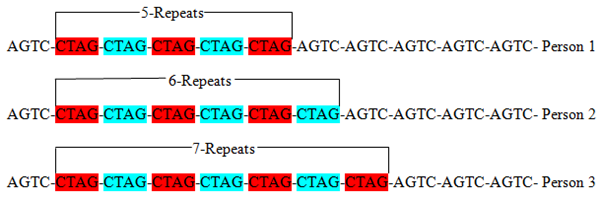
Figure 3 Showing tetra-nucleotide (CTAG) STRs of different lengths at the same locus.3
In the forensic settings, STRs are suitable marker for identification purpose owing to the following factors; suitability for easy amplification by polymerase chain reaction (PCR), its variable nature of short repeat sequences among individuals, small in size which gives it the advantage for degraded samples, low mutation rate and high power of discrimination.4,30,31 In light of this, the introduction of an amplification technology linked to STRs method of DNA profiling resulted to the availability of appropriate vigorous systems for the establishment of an effective and efficient DNA database.32-34 Since the enactment of a comprehensive legislation in the UK in 1995 for the establishing of the first DNA database capable of storing the personal DNA profiles alongside profiles obtained from crime scene biological samples using the STRs as a gold standard, tremendous successes have been recorded in providing compelling evidences for the administration of justice.3,17 Following a long period of operation, the National DNA Index System (NDIS) has been demonstrated to increase in relevance and size in conjunction with STR DNA technology,3,35 and giving the study of Budowle et al. (1998), in 1997, 13 core STR loci namely: CSF1PO, FGA, TH01, TPOX, vWA, D3S1358, D5S818, D7S820, D8S1179, D13S317, D16S539, D18S51, and D21S11 were in use, however, in January 2017, the number of loci for new CODIS profiles was increased to 21 including the sex marker amelogen into enhance the discriminating power.15,24,35,36 STRs technology continues to be a strong workhouse for forensic DNA profiling identification and when the recommended loci are in use, the average random match probability for unrelated persons is rarer that one in a trillion.37
Variable number tandem repeat (VNTR)
Variable number tandem repeat (VNTRs) is also known as the minisatellite consisting of core repeated sequences of greater than six (> 6) mostly sequences of about (20-100 bases) repeated between 0 to 30 times. Minisatellites repeats are found in the genome organized as tandem repeats flanked by segments of non-repetitive sequences.7 They are found to be located on many chromosomes, and the number of tandem sequences repeats in a certain locus are vastly variable among unrelated persons.8 Additionally, each minisatellite is composed of a core repeated sequences, and some of the core sequences are found to be similar to all individual, however, the variation is in the occurrence of core repeated sequence at a specific locus,38 as revealed in figure 4. Therefore, two individuals may have the number of repeat sequences at one locus, but the possibility of sharing the same number of repeat sequence at multiple loci is significantly negligent.39
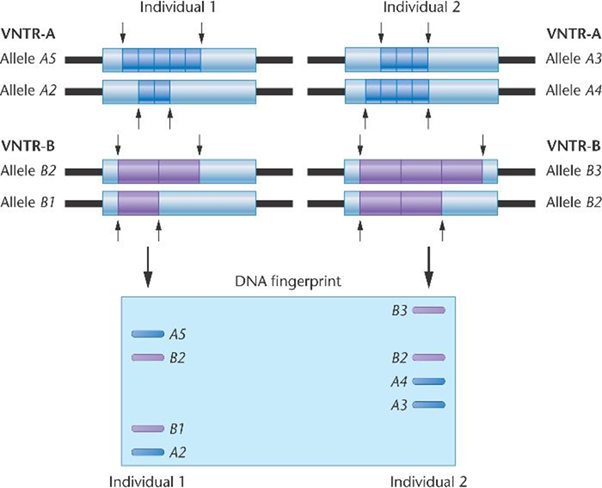
Figure 4 Variable number tandem repeat for identification.40
Figure 4 depicts a variation in the core repeat sequence of two individual at a various locus. It is seen that at locus 1 labelled (VNTR-A), there was five repeat of a given tandem sequence for allele A5, and two for repeat sequence for allele A2 for individual 1. Conversely, for individual 2, three repeat sequence for allele A3, and four repeat sequence for allele A4. The variation is also observed for the second locus labelled (VNTR-B). Therefore, efficiency of VNTR marker is a great resource for identification purpose.
Restriction fragment length polymorphisms (RFLP)
Restriction fragment length polymorphism (RFLP) is molecular marker that utilizes the enzyme restriction endonuclease which cuts DNA strands at a specific site called the restriction site to produce fragments of DNA or restriction fragments of defined length.9 Practically and in general, the restriction endonuclease enzyme is incubated with the DNA molecules for digestion, the digested product is then size separated using electrophoretic technique either agarose or acrylamide gel electrophoresis as revealed in figure 5.41 Summarily, the steps for DNA fingerprinting using RFLP are as follows;
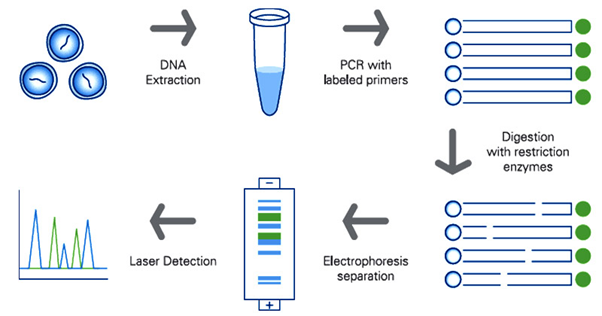
Figure 5 Steps for DNA profiling using restriction length polymorphism.42
In the context of forensics, RFLP was a prominent resource for identification purposes before it was later replaced with a STR marker, a simpler and sensitive technique.8,41 RFLP individual variation is differentiated by the analysis of patterns of DNA cleaved by restriction enzymes. In this light, if there is a variation in the distance between the point of cleavage of a specific restriction enzyme, the fragment length produced will vary when the DNA is digested with a restriction endonuclease.7Owing to the variation in DNA fragment at a specific restriction sites, it is still in use for identification purposes particularly in paternity cases.
DNA profiling markers continues to be a great resource for identification purposes, and its technology for DNA fingerprinting in the context of forensic differs in their ability in differentiating two persons, and also in the rapidity and sensitivity of obtaining the result. Since, the establishment of the first National DNA database in the UK, and the validated technology of short tandem repeat, short tandem repeat markers has become the gold standard for forensic DNA fingerprinting, although variable number tandem repeats and restriction fragment length polymorphism still remain in use, particularly for paternity cases and also for prokaryotic gene mapping and sequencing. We also consider this article to be an important resource for budding forensic geneticist.
None.
Authors declare that there is no conflict of interest.

©2020 Nwawuba, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.