eISSN: 2379-6367


Research Article Volume 6 Issue 6
1Molecular Biology Laboratory, Butantan Institute, Brazil
2Alchemy-Innovation, Research & Development, CIETEC-IPEN-University of Sao Paulo, Brazil
3Immunochemistry Laboratory, Butantan Institute, Brazil
Correspondence: Kerly Fernanda Mesquita Pasqualoto, Alchemy-Innovation, Research & Development, Prof. Lineu Prestes Ave. 2242, Room 113, CIETEC-IPEN-University of Sao Paulo, Sao Paulo, SP-05508-000, Brazil
Received: October 25, 2018 | Published: November 9, 2018
Citation: Pasqualoto KFM, Silva BAVG, Kodama RT, et al. Exploring the C-domain inhibition of angiotensin converting enzyme through novel bradykinin potentiating peptides. Pharm Pharmacol Int J. 2018;6(6):408-416. DOI: 10.15406/ppij.2018.06.00210
Background: Proline-rich oligopeptides (PRO) presenting antihypertensive effects have been found in snake venoms. They can be selective to the C-terminal domain of angiotensin converting enzyme (ACE) binding site and quite potent competitive inhibitors regarding the angiotensin I cleavage, as well. The main structural features of PRO ligands comprise: a pyroglutamyl residue at the N-terminal portion, a high content of proline residues, and the tripeptide Ile-Pro-Pro at the C-terminal moiety.
Objective: A set of eight PRO compounds from venom of Bitis and Bothrops genus was investigated, herein, using molecular docking and structure-property approaches to explore the ACE C-terminal domain.
Method: Eight PRO compounds were selected according to the respective inhibition constant value against the ACE enzyme. The coordinates of the human ACE/BPP complex were retrieved from Protein Data Bank (PBD ID 4APJ; resolution at 2.60 Å) and used as reference to perform the molecular docking simulations, using CLC Drug Discovery Workbench 2.4 software. The three-dimensional (3D) structure of the ligand in the 4APJ complex was considered as starting geometry to build up the eight PRO molecular models.
Results: The compound PRO8 presented favorable calculated binding affinity, but compounds containing additional amino acid residues at the C-terminal moiety, such as PRO2 and PRO4, have showed poor docking score values, meaning the ligand-enzyme complex formation was energetically unfavorable.
Perspectives: Since BPPs have been reported as multi-target compounds, the novel promising compound, PRO8, from Bitis nasicornis venom, can be optimized and drive the rational design of antihypertensive drug candidates considering the two pathways involved in BPPs’ hypotensive effects.
Keywords: ACE inhibitors, bradykinin potentiating peptides, molecular docking simulations, structure-property relationships, snake venoms, rational drug design
PRO, Proline-rich oligopeptides; BPP, bradykinin-potentiating peptides; ACE, angiotensin converting enzyme; RAS, renin-angiotensin system; BK, bradykinin; PDB, protein data bank; ID, identification; 3D, three-dimensional; EP, electrostatic potential; CHELPG, charges from electrostatic potential using a grid based method; DFT, density functional theory; B3LYP, Becke 3-Parameter (Exchange); Lee, Yang and Parr method, RMSD, root mean square deviation; MEP, map of electrostatic potential; V, molecular volume
Toxins are complex mixtures of bioactive substances which may target many physiological processes. They are highly selective and potent compounds, and frequently can be used as lead compounds in the drug development process. Proline-rich oligopeptides (PRO) from Bothrops jararaca venom, for instance, have presented interesting effects on the cardiovascular system, providing information for developing anti-hypertensive drugs.1‒4 Captopril is a classic example of drug development based on animal venoms. It was the first angiotensin converting enzyme (ACE) inhibitor developed using the rational drug design approach, where novel drug candidates are conceived primarily considering the pathophysiology mechanisms, or the biochemical pathways, involved in a target disease process.5,6 When the structural information regarding the target biomacromolecule (enzyme, receptor, DNA, etc.) and its related ligands (natural or synthetic) are available (X-ray diffraction or NMR), computer-aided molecular design approaches, such as structure-based drug design (SBDD) can also be applied.5‒7 The SBDD strategy is well established and has been extensively used in drug discovery and development processes by the pharmaceutical companies.5‒8 Molecular docking is an important step involving in SBDD widely applied on hit identification. Docking procedure combined to a scoring function allows identification of several compounds and ranking the best ligands considering three-dimensional complementarity between ligand and target.9,10
The renin-angiotensin system (RAS) (Figure 1) is involved in the regulation of blood pressure, and the key enzyme in this process is the ACE, which is responsible for inactivating bradykinin (BK), decreasing the hypotensive effect, and concurrently is responsible for converting angiotensin I to angiotensin II, which presents a vasopressor effect.11,12 In this regard, ACE inhibitors can display antihypertensive effects either by blocking the formation of angiotensin II or by enhancing the BK hypotensive effect.13 Concerning the structural analysis of somatic ACE, the extracellular region of the protein has two homologous catalytic domains (N- and C-terminus). Both domains contribute to the BK degradation. The selective inhibition of ACE C-domain, however, prevents induced vasoconstriction by angiotensin I. The C-domain is primarily responsible for the conversion of angiotensin I into angiotensin II14 and, consequently, can be sufficient to decrease blood pressure.15‒17 High concentration of BK may cause angioedema as side effect, though.18 Therefore, the structural information regarding the ACE C-domain should be considered for designing more promising and selective drug candidates.
Even though there are antihypertensive drugs available on the market, the blood pressure has been controlled only in a half of cases,19 since many of those drugs are nonspecific on the RAS. More specific inhibitors, providing better effect on the blood pressure regulation as well as better safety and tolerability on the pharmacological profile have been considered as therapeutic alternatives.20‒22 Proline-rich oligopeptides (PRO), or bradykinin-potentiating peptides (BPP), which can be found in snake venom, were the first naturally ACE inhibitors described, and have been reported as presenting antihypertensive effects related to the C-domain inhibition.13,23,24 PRO compounds have been characterized by containing
Kodama et al.25 have investigated the presence of ACE modulators in venoms of different species from Bitis genus, and identified novel PRO compounds acting as inhibitors preferentially on the C-domain of ACE binding site. Based on their findings, the binding mode of eight inhibitors were investigated applying molecular docking simulations to
Moreover, the structure-property relationships were also assessed to better understand the amino acid substitution pattern among the investigated compounds. In this regard, the map of electrostatic potential was calculated onto the molecular surface of each PRO compound to verify changes regarding the amino acid substitution pattern. The findings may help to drive the structure-based drug design of novel compounds (peptidomimetics) regarding the antihypertensive response.
Molecular docking findings
The ACE inhibitors binding mode, in general, depends on the establishment of a molecular interaction with a zinc ion, which is the enzyme’s cofactor. ACE inhibitors use to have a chemical portion in its structure capable of forming a coordinate bond with a zinc ion to provide the biological response.11,12 Masuyer et al.26 however, have elucidated the molecular interactions of natural peptide inhibitors (BPPs) regarding the ACE C-terminal site. They have revealed, for the first time, the detailed molecular interactions in a zinc independent manner considering the structure of the complex ACE-BPP were the interaction between water molecule and proline residue on C-terminal on BPPb was described as responsible for losing coordination with zinc ion from ACE binding site. Even with Mansuyer and co-workes results, we carried out docking procedure in the presence of the zinc ion. Our findings suggests that residue 8 at PRO1, PRO3, PRO5, PRO6 and PRO8 could interact with zinc, was observed distances between residue 8 at each PRO and zinc range 2.957 to 3.268Å. PRO7 also can interact with zinc (distance at 3.064Å) but through residue 9. Zinc ion was observed between proline residues at position 9 and 10 from PRO2 and PRO4 but not indicating possibility of interaction. Zinc binding motif HEXXH27 correspond to H383, E384, M385 and G386 residues on human ACE/BPP complex selected for this study (PDB code 4APJ, resolution at 2.60 Å).26
The eight PRO compounds were docked in the ACE binding site using as reference the 4APJ ligand,26 which presents eleven amino acid residues (<EGLPPRPKIPP). Of note, the last seven residues in the 4APJ ligand’s sequence are identical to those in the PRO1 compound’s sequence. The re-docking score was energetically favorable (negative energy value; -134.07 kcal.mol-1) and the root mean square deviation (RMSD) value was less than 2Å28 indicating that the optimum conditions to perform the molecular docking simulations had been properly defined. The energy score (hydrogen bond interactions, steric interactions, ligand conformational penalty), RMSD (Å), and experimental Ki (µM)25 values for each ACE-BPP complex are listed in Table 1.
|
Ligand |
Kia (nM) |
Total score (kcal.mol-1) |
RMSD (Å) |
Hydrogen bond interactions score (kcal.mol-1) |
Steric interactions score (kcal.mol-1) |
Ligand conformation penalty (kcal.mol-1) |
|
4APJb |
- |
-134,07 |
0.16 |
-26.66 |
-125.75 |
18.35 |
|
PRO8 |
250 |
-89.35 |
0.06 |
-33.98 |
-107.73 |
52.36 |
|
PRO3 |
280 |
-83.84 |
0.06 |
-40.5 |
-105.41 |
62.07 |
|
PRO1 |
480 |
-80.58 |
0.03 |
-34.26 |
-97.61 |
51.29 |
|
PRO5 |
200 |
-74.72 |
0.01 |
-31.21 |
-104.76 |
61.26 |
|
PRO6 |
830 |
-62.03 |
0.02 |
-31.32 |
-96.23 |
65.52 |
|
PRO7 |
470 |
-20.41 |
0.13 |
-26.76 |
-103.12 |
109.47 |
|
PRO4 |
>100,000 |
266.82 |
2.36 |
-5.1 |
180.79 |
91.13 |
|
PRO2 |
>100,000 |
380.45 |
0.11 |
-6.93 |
291.04 |
96.34 |
The calculated binding affinity values of compounds PRO1-8 ranged from -89.35 to 380.45kcal/mol and the RMSD values varied from 0.01 to 2.36Å (Table 1). According to the scoring index, six out of eight compounds presented favorable energy values (negative values) concerning the enzyme-ligand complex formation. The energy values are related to the calculated binding affinity of the compounds regarding the ACE interaction site, as aforementioned. The two compounds, PRO4 and PRO2, which presented higher energy values (positive values), also showed higher experimental Ki values (>100µM), validating the in silico approach. Of note, the ten first amino acid residues of compounds PRO4 and PRO2 correspond to the sequences of compounds PRO3 and PRO1, respectively. However, compounds PRO4 and PRO2 have two more amino acid residues, Met11 (apolar uncharged) and Lys12 (polar positively charged), after the tripeptide Ile-Pro-Pro at the C-terminal moiety, which were quite likely responsible for impairing their accommodation in the ACE binding site reducing significantly their binding affinity.
Concerning the experimental inhibitory activity (Ki values; µM25), the compounds could be divided into three groups: more active (PRO5, PRO8, PRO3), Ki values from 200 to 280µM; moderately active (PRO7, PRO1, PRO6), Ki values from 470 to 830µM; and, less active (PRO2, PRO4), Ki values higher than 100µM. Based on the total docking score, some compounds could be classified differently, though. PRO1, for instance, which presented more negative energy value than PRO5 would be also more active instead of moderately active. The total docking score is composed by the sum of energy contributions from hydrogen bond interactions, steric interactions and ligand conformation penalty (Table 1). Among the limitations of molecular docking approach are the size and structural freedom degrees (flexibility) of ligands. In this regard, constrains must be considered to properly perform docking simulations. Herein, the compounds were treated as rigid molecules and the ligand conformation penalty contribution reflects that constrain. When the ligand conformation penalty contribution is disregarded from the total docking score, the compounds’ classification based on the values of calculated binding affinity is more consistent with the experimental inhibitory data.
Regarding the amino acid substitution pattern, the difference between the two more active compounds, PRO5 (<ENWPHPQIPP) and PRO3 (<ENWPRPQIPP), relies on the fifth residue, His5 and Arg5, respectively. Both residues are polar positively charged sharing similar molecular properties, though. The novel active compound, PRO8 (<ENWPRPKVPP), presents also an arginine residue at fifth position, similarly to PRO3, but it differs from both active compounds mainly at the seventh position. Instead of Gln7, a polar uncharged residue, it has a polar positively charged residue (Lys7). The eighth residue, which is part of the tripeptide (Ile-Pro-Pro) at the C-terminal moiety, maintains the hydrophobic feature to all three compounds (Ile8 or Val8). According Cotton and co-workers (2002),13 the fourth position on PROs and sequence signature IPP drives potency and selectivity for ACE C-domain. PRO5 correspond to BPP2 (<ENWPHPQIPP), contains a glutamine at position 4 and was described as a high selective C-domain compound compared to PROs with lysine at fourth positon.13
Comparing the sequences of moderately active compounds, PRO1 and PRO6, to those of most active compounds, PRO8 and PRO5, the difference relies, respectively, on the amino acid substitution at the eighth position (Ile8 or Val8). Even though sharing hydrophobic properties, Ile is bulkier than Val residue. The steric interactions score values found for the compounds PRO8 (more active) and PRO1 (moderately active), for instance, were -107.73 and -97.61kcal/mol, respectively, pointing out that the Val residue would provide interactions more energetically favorable into the ACE binding site regarding the amino acid pattern <ENWPRPKXPP (X=Val). On the other hand, for the compounds PRO5 (more active) and PRO6 (moderately active), the steric interactions score values were -104.76 and -96.23kcal/mol, respectively, emphasizing the Ile residue at the C-terminal moiety as more energetically favorable for the amino acid pattern <ENWPHPQXPP (X=Ile) concerning the interactions established into the ACE binding site.
Interestingly, the compound PRO7 (moderately active) has a very different amino acid substitution pattern, despite the conserved tripeptide Ile-Pro-Pro at the C-terminal moiety (<EWQRPGPEIPP). Regarding the seven last residues, there is a glycine (polar; non-substituted amino acid) at the fifth position instead a positively charged residue. In addition, it has a negatively charged residue at the seventh position instead a polar uncharged (Q) or a positively charged (K) amino acid. Concerning the N-terminal moiety, there are three (WQR; W=hydrophobic, Q=polar uncharged, R=polar positively charged) instead of two (NW; N=polar uncharged, W=hydrophobic) residues after the pyroglutamyl residue. Despite all molecular changes, the compound PRO7 has presented an energetically favorable value (negative; -103.12kcal/mol) for the steric interactions score contribution. However, it had the highest value for the ligand conformation penalty contribution (109.47kcal/mol), and instead of being classified as the fourth compound regarding its experimental inhibitory activity (470µM), it was placed at the sixth position (total score = -20.41kcal/mol; Table 1).
According to the data previously reported by Kodama et al.25 the inhibitors experimentally more specific to the C domain of somatic ACE were the compounds PRO3 (<ENWPRPQIPP), PRO5 (<ENWPHPQIPP), PRO6 (<ENWPHPQVPP), and PRO8 (<ENWPRPKVPP). Of note, the three first amino acid residues are the same to those ligands. The differences among the sequences rely mainly on the residues at fifth, seventh, and eighth positions, as aforementioned. Furthermore, regarding the findings from molecular docking simulations, the compounds PRO3, PRO5, PRO6, and PRO8, have established, respectively, twelve, eight, eight, and eleven hydrogen bonding interactions into the ACE binding site (Table 3). Hydrogen bonding interactions involving Tyr62 and the first (PRO5, PRO6, PRO8) and second (PRO3) residues nearby the N-terminal region were found only for more active PRO compounds. The list of amino acid residues into the ACE-binding site that interact with each PRO compound is presented in Table 2. The molecular interactions between the compounds PRO3, PRO5, PRO6, PRO8 and the complementary amino acid residues in the ACE binding site are shown in Figure 2.
More active compounds |
Moderately active compounds |
Less active compounds |
|||||
PRO5 |
PRO8 |
PRO3 |
PRO1 |
PRO7 |
PRO6 |
PRO4 |
PRO2 |
Tyr23 (1) |
Tyr23 (2) |
Tyr23 (1) |
Arg85 (1) |
Tyr12 (3) |
Tyr23 (1) |
Thr53 (2) |
Thr53 (3) |
Gln242 (1) |
Gln242 (1) |
Asn46 (1) |
Gln242 (1) |
Thr53 (1) |
Gln242 (1) |
Cys313 (1) |
Tyr321 (1) |
Ala317 (1) |
Ala317 (1) |
Gln242 (1) |
Ala317(1) |
Gln242 (1) |
Ala317 (1) |
Tyr321 (1) |
Tyr480 (2) |
Tyr321 (1) |
Tyr321 (1) |
Ala317 (2) |
Tyr321 (1) |
Ala317 (2) |
Tyr321 (1) |
Cys331 (1) |
Tyr12 |
Lys468 (1) |
Glu364 (2) |
Tyr321 (1) |
Glu364 (1) |
Tyr321 (1) |
Lys468 (1) |
Tyr480 (1) |
Ser16 |
Tyr477 (1) |
Lys468 (1) |
His348 (1) |
Lys468 (1) |
Lys468 (1) |
Tyr477 (1) |
Tyr12 |
Val19 |
Tyr480 (2) |
Tyr477 (1) |
Lys468 (1) |
Ser474 (1) |
Tyr477 (1) |
Tyr480 (2) |
Tyr23 |
Tyr23 |
Tyr12 |
Tyr480 (2) |
His471 (1) |
Tyr477 (1) |
Tyr480 (1) |
Tyr12 |
Ala50 |
Ala24 |
Val19 |
Tyr12 |
Ser474 (1) |
Tyr480 (2) |
Ser16 |
Val19 |
His52 |
Asn46 |
Ala24 |
Ala24 |
Tyr477 (1) |
Tyr12 |
Tyr23 |
Ala24 |
Leu54 |
Gln48 |
Asn46 |
Asn46 |
Tyr480 (1) |
Ser16 |
Ala24 |
Asn46 |
Tyr56 |
Ala50 |
Ala50 |
Ala50 |
Leu83 |
Val19 |
His52 |
Ala50 |
Gly57 |
His52 |
Val80 |
Val80 |
Ala86 |
Tyr23 |
Tyr56 |
Val80 |
Val80 |
Val80 |
Asn97 |
Gln81 |
Tyr96 |
Ala24 |
Gly57 |
Asn97 |
Gln81 |
Leu83 |
Tyr355 |
Asn97 |
Glu104 |
Ala50 |
Val80 |
Tyr355 |
Leu83 |
Ala86 |
His371 |
Tyr355 |
Phe414 |
Val80 |
Gln81 |
Phe414 |
Ala86 |
Tyr96 |
Phe414 |
Gly365 |
Phe469 |
Leu83 |
Leu83 |
Phe469 |
Ala87 |
Leu100 |
Phe469 |
His371 |
Ser473 |
Ala86 |
Ala86 |
His470 |
Glu123 |
Glu123 |
His470 |
Phe414 |
Val475 |
Tyr355 |
Gln330 |
Val475 |
Gln242 |
Trp181 |
Val475 |
Phe469 |
Pro476 |
His371 |
Ala361 |
Lys329 |
Gln242 |
|
His470 |
Phe414 |
Leu362 |
Gln330 |
Lys329 |
|||
Val475 |
Phe469 |
His470 |
Thr332 |
Gln330 |
|||
His470 |
His371 |
Thr333 |
Glu337 |
||||
Ser473 |
Phe414 |
Asp338 |
Asp338 |
||||
Val475 |
Phe469 |
Val340 |
Tyr355 |
||||
Pro476 |
Val475 |
Tyr355 |
Ala361 |
||||
Ala361 |
His371 |
||||||
Val375 |
Phe469 |
||||||
Asp376 |
His470 |
||||||
Asp410 |
Ser473 |
||||||
Phe414 |
Ser474 |
||||||
Lys468 |
Val475 |
||||||
His470 |
Pro476 |
||||||
His471 |
Ser483 |
||||||
Ser473 |
|||||||
Ser474 |
|||||||
Tyr477 |
|||||||
|
|
|
|
|
|
Ser483 |
|
Table 2 List of the amino acid residues which establish interactions with the PRO compounds into the ACE-binding site, according to the findings from molecular docking simulations (CLC Drug Discovery Workbench Software33)
The residues in the ACE-binding site that establish hydrogen bonding interactions are in bold letters followed by the number of H bonds established between parentheses

Figure 2 Complexes enzyme-ligands from molecular docking simulations displaying the ACE C-domain and ligands considered as experimentally more specific: (a’) schematic representation of the complex ACE-PRO3 (ligand in blue; hydrogen atoms are hidden); (a’’) molecular interactions between the ligand PRO3 (stick model; color by element) and the complementary residues in the ACE binding site; (b’) schematic representation of the complex PRO5-ACE (ligand in purple; hydrogen atoms are hidden); (b’’) molecular interactions between the ligand PRO5 (stick model; color by element) and the complementary residues in the ACE binding site; (c’) schematic representation of the complex ACE-PRO6 (ligand in dark green; hydrogen atoms are hidden); (c’’) molecular interactions between the ligand PRO6 (stick model; color by element) and the complementary residues in the ACE binding site; (d’) schematic representation of the complex ACE-PRO8 (ligand in cyan; hydrogen atoms are hidden); (d’’) molecular interactions between the ligand PRO8 (stick model; color by element) and the complementary residues in the ACE binding site. The interatomic distances are less than 3.5 Å. Alpha-helices are shown as red cylinders and beta-sheets are presented as cyan flat arrows (CLC Drug Discovery Workbench Software,36 Discovery Studio Visualizer43).
Interestingly, molecular docking simulations have provided the identification of common amino acid residues, into the ACE binding site, which can establish interactions with the PRO compounds, such as: Trp59, Tyr62, Asn66, Ile88, Thr92, Lys118, Asp121, Glu123, Arg124, Gln281, His353, Ala354, Ser355, Ala356, Trp357, Asp358, Val380, His383, Glu384, His387, Phe391, Val399, Arg402, Glu403, Glu411, Arg522, Tyr523, and Phe527. Moreover, hydrogen bonding interactions between the C-terminal region of each ligand and two water molecules (Wat2081 and Wat2089) into the binding site were also observed. For the PRO2 compound, only one water molecule (Wat2089) established interactions with the C-terminal moiety, though. In addition, the Tyr360 and Tyr523 residues seem to be important in the ligands’ binding mode and affinity. The ninth and fourth amino acid residues in the sequence of PRO compounds established hydrogen bonding interactions, correspondingly, with Tyr523 and Tyr360. For the compounds PRO7 and PRO2, though, the amino acid residues involved in those hydrogen bonding interactions are placed at the fifth and seventh positions, respectively. Other binding site residues participating in hydrogen bonding interactions with the more active PRO compounds are: Gln281, Ala356, Lys511, and Tyr520. The Gln281, Lys511 and Tyr520 residues interact with the amino acid residues at the tenth position in the sequence of more active PROs whereas the Ala356 residue interacts with the amino acid residues placed at the seventh position in the sequence (except to compound PRO7, where Ala356 interacts with the amino acid residue at the eighth position).
At molecular level, the experimentally less active compounds, PRO4 and PRO2, have presented distinct binding mode establishing a larger number of non-bonded interactions in the ACE binding site (Table 2). However, less amino acid residues into the ACE binding site were participating in hydrogen bonding interactions with those ligands: five residues to PRO4 (Thr92, Cys352, Tyr360, Cys370, Tyr523; six hydrogen bonding interactions) and three residues to PRO2 (Thr92, Tyr360, Tyr523; six hydrogen bonding interactions). Furthermore, the amino acid residues in the binding site nearby the Met11 and Lys12 residues of ligands PRO4 and PRO2 were, respectively, Lys368, Gln369, Cys370, Thr371, Thr372, Asp377, Val379, Val414, Asp415, Asp453, for PRO4; and Lys368, Gln369, Glu376, Asp377 (Table 2). Therefore, the different accommodation in the ACE binding site provided higher total docking score values (positive values; energetically unfavorable), which are in accordance to the lack of experimental inhibitory activity (Ki values>100µM). The findings have pointed out that the residues after IPP or VPP signature moiety changes the usual interaction points, for instance highly impairing the accommodation of the ligand at the C domain site. Thus, changing the usual interaction points would not be a proper strategy for designing novel more specific ACE inhibitors. The chemical structure of compounds is responsible for their physicochemical and reactivity properties reflecting directly on the molecular recognition process and, consequently, on the formation of the ligand-target complex, which provides the biological response.5,6,29 Beside decreasing the main chain conformational freedom, the proline residues may constrain torsion angles mostly keeping peptides in beta-sheet conformation,30,31 for instance.
Structure-property findings
The map of electrostatic potential (MEP), which is an electronic molecular property, was calculated onto the molecular surface of each compound to visualize the ligands’ amino acid substitution patterns (side chains) in terms of electronic density distribution. Since the molecular surface is related to the compound’s stereochemistry (spatial distribution of the functional groups), it translates the compound’s molecular shape. The electrostatic potential and molecular shape can be considered the two primary properties in the molecular recognition process.32 The MEPs calculated to the PRO compounds experimentally more specific regarding the ACE C-domain (PRO3, PRO5, PRO6, PRO8) are shown in Figure 3.
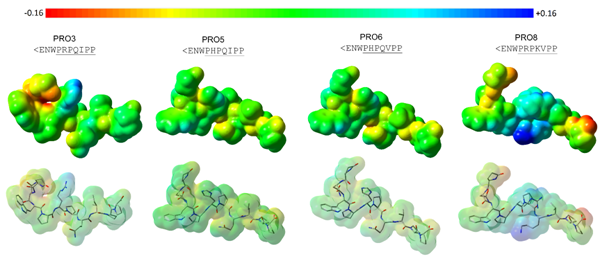
Figure 3 Maps of electrostatic potential (MPEs) calculated onto the molecular surface of compounds considered as experimentally more specific to the ACE C-domain (PRO3, PRO5, PRO6, and PRO8). Regarding the color range, higher electronic density distribution regions are displayed as intense red color (-0.160) and lower electronic density distribution regions are shown in intense blue color (+0.160) (Gaussian 03W;41 GaussView 0542).
Focusing on the molecular surface (shape), the compounds PRO5 (<ENWPHPQIPP) and PRO6 (<ENWPHPQVPP) are quite similar. However, they differ from one another only by the residue at the eighth position (Ile8 and Val8), as aforementioned. Both residues share similar molecular properties (hydrophobic residues), but Ile is bulkier than Val. The calculated molecular volume (V) values for compounds PRO5 and PRO6 were 1081.19 and 1065.83Å3, respectively. The bulkier residue at the C-terminal moiety could be responsible for the difference in inhibitory activity. Observing the MEPs, the electronic density distribution on the eighth residue region is more neutral (green color), according to the color scheme shown in Figure 3.
The compounds PRO8 (<ENWPRPKVPP) and PRO3 (<ENWPRPQIPP) presented different molecular shape nearby the residues at fifth (Arg5 and His5) to seventh (Lys7 and Gln7) positions when compared to the compound PRO5 (<ENWPHPQIPP). Arginine is bulkier than histidine, and lysine is bulkier than glutamine, as well. The calculated molecular volume values found for PRO8 and PRO3 were, respectively, 1160.43 and 1113.85Å3. Both compounds are bulkier than compound PRO5 (V=1081.19Å3). Regardless the molecular volume difference, the three compounds have polar positively charged residues at fifth position. But, the compound PRO8 has a lysine residue (polar positively charged) at seventh position instead of glutamine (polar uncharged). Concerning the MEPs, the presence of two positively charged residues in the sequence of PRO8 has provided a more intense blue color on the portion of molecular surface nearby the fifth to seventh residues, reinforcing a lower electronic density distribution (more positive region).
Of note, the sequence of compound PRO7 (<EWQRPGPEIPP; moderately active) comprises the most different amino acid substitution pattern. It has one additional amino acid residue in the N-terminal portion (Arg4), and the Trp residue (hydrophobic and bulky) is placed at the second position, not at the third, providing changes in the compound’s molecular shape. Also, the Gln residue (at the third position) is polar uncharged, but bulkier than Asn (polar uncharged), which is placed at the second position in the sequence of the others PRO compounds. The presence of an arginine (positively charged) at the fourth position provides not only changes in the molecular shape (V = 1169.17Å3) but also in the electronic density distribution on the initial portion of the compound (Figure 4). Regarding the last seven residues (underlined sequence), the main differences rely on the Gly6 (not substituted amino acid residue) and Glu8 (polar negatively charged) residues, which have also provided changes in both, molecular shape and electronic density distribution (MEP) of compound PRO7, as shown in Figure 4.
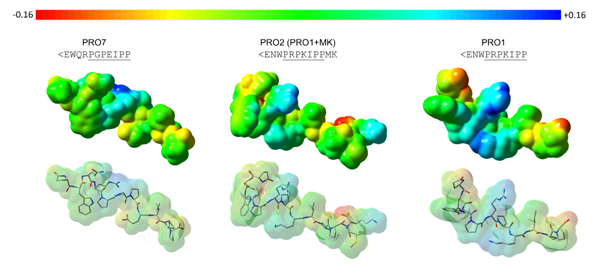
Figure 4 Maps of electrostatic potential (MPEs) calculated onto the molecular surface of compounds PRO7 (moderately active), PRO2 (less active), and PRO1 (active). According to the color range, higher electronic density distribution regions are displayed as intense red color (-0.160) and lower electronic density distribution regions are shown in intense blue color (+0.160) (Gaussian 03W;41 GaussView 0542).
As aforementioned, the compound PRO2 (<ENWPRPKIPPMK; less active) has the same amino acid sequence as compound PRO1 (<ENWPRPKIPP; moderately active) plus two more residues at the C-terminal portion, Met11 (apolar uncharged) and Lys12 (polar positively charged). By adding these two residues, the molecular shape and electronic density distribution have changed (Figure 4), reflecting on the binding mode and inhibitory activity of the two compounds. The calculated molecular volume values found for PRO2 and PRO1 were, respectively, 1375.2Å3 and 1126.78Å3.
Despite of being studied for decades, the ACE enzyme remains as the primary molecular target for developing more selective and specific antihypertensive drugs, since it is the key enzyme of the renin-angiotensin system. PROs may inhibit the ACE C-terminal site, and studies have also shown its effects on nitric oxide release at vascular endothelium, suggesting that the hypotensive effect could be due to the action on two pathways.33 In this regard, PROs can be considered as multi-target compounds, meaning they have affinity by different targets involved in the same dysfunction. Herein, we have exploited the binding mode (calculated affinity) and the structure-property relationships of a set of PRO compounds from snake venoms. A novel compound, PRO8, has presented the best calculated affinity by the ACE binding site. The findings from molecular docking simulations and calculated molecular properties have allowed the proposition of PRO sequence (structural) requirements regarding the establishment of more specific interactions into the ACE C-domain (Figure 5). Besides the molecular interactions shown to the crystallographic ligand 4APJ26 (Gln281, Ala356, Lys511, Tyr520, and Tyr523), which were also observed herein for the compounds experimentally more specific to the C-domain, our findings have indicated the residues Tyr62, Tyr360, Phe457, Phe512, and Val518, as important interaction sites for developing more specific ACE C-domain inhibitors. The novel compound, PRO8, can be considered as a new hit to drive the rational designing of anti-hypertensive drug candidates taken into account, for instance, two pathways such as renin-angiotensin system and nitric oxide release. More specific drugs acting on the ACE enzyme may better control the arterial pressure in patients, but in order to make significant differences in therapy, multi-target drugs have been shown more promising.

Figure 5 Structural requirements for the PRO compounds regarding the establishment of more specific interactions into the ACE C-domain. The important complementary residues into the binding site are displayed as well as the respective amino acid positions in the PRO sequence involved in molecular interactions more specific to the C-domain. Hydrogen bonding interactions, which were observed to all investigated PRO compounds, are indicated as green dashed lines (Tyr23, Gln242, Lys468, Tyr477), and those observed only to more specific C-domain inhibitors are highlighted as blue dashed lines (Tyr321, Tyr480); molecular interactions involved the amino acid residue backbone (Ala317) are displayed as blue dotted lines; the Phe414, Phe469, and Val475 residues establish non-bonded hydrophobic interactions indicated by double gray semi-curves.
Eight PRO compounds were selected according to the respective inhibition constant (Ki µM) value against the ACE enzyme. Of note, all compounds were tested under the same pharmacological protocol using the somatic ACE, which presents the two homologous catalytic domains.25 Except to the compounds PRO3 and PRO5, which have already been found in Bothrops jararaca venom,34 the others were isolated and experimentally tested for the first time by Kodama et al.25 The BPP code, amino acid sequence, and organism source are listed in Table 3. The coordinates of the human ACE/BPP complex were retrieved from Protein Data Bank,35 PBD ID 4APJ (resolution at 2.60Å),26 and used as reference to perform the molecular docking simulations, using CLC Drug Discovery Workbench 2.4 software.36 The three-dimensional (3D) structure of the ligand in the 4APJ complex was considered as starting geometry to build up the eight PRO molecular models, PRO1-8.
Compound code |
Sequence |
Organism source |
PRO1 |
<ENWPRPKIPP |
Bitis nasicornis |
PRO2 |
<ENWPRPKIPPMK |
Bitis nasicornis |
PRO3 |
<ENWPRPQIPP |
Bothrops jararaca |
PRO4 |
<ENWPRPQIPPMK |
Bitis nasicornis |
PRO5 |
<ENWPHPQIPP |
Bothrops jararaca |
PRO6 |
<ENWPHPQVPP |
Bitis gabonica rhinoceros |
PRO7 |
<EWQRPGPEIPP |
Bitis gabonica gabonica |
PRO8 |
<ENWPRPKVPP |
Bitis nasicornis |
Table 3 PRO code, amino acid sequence, and organism source regarding the set of compounds investigated
<E corresponds to the pyroglutamyl residue.
Re-docking of the 4APJ ligand (<EGLPPRPKIPP) was carried out in order to establish the optimum conditions for computing the BPPs binding affinity values. The conditions used to perform the molecular docking simulations were the following:
Not only the root-mean square deviation (RMSD) values concerning the atomic positions of the 4APJ ligand and each PRO compound, but also the score by PLANTSPLP method38 from docking procedure were employed as evaluation criteria. Of note, the binding mode of each ligand in the protein binding pocket is related to a score value. The score, herein, mimics the potential energy change when the target protein and ligand come together, meaning that a very negative score corresponds to a strong binding whereas a less negative, or even positive, score value corresponds to a weak or non-existing binding affinity. The total score value comprises the following types of contribution: hydrogen bond score, metal interaction score, steric interaction score, and ligand conformation penalty score. Concerning the last contribution, it scores the complementarity between the binding site and ligand by rewarding and punishing different types of heavy atom contacts having inter-atom distance less than 5.5Å.36
Furthermore, the electrostatic potential (EP) property of each ligand (BPP) was calculated to visualize the changes in electronic density distribution concerning the amino acid substitution patterns. The charges from electrostatic potential using a grid based method (CHELPG)39 were calculated for each 3D molecular model (PRO1-8) employing the density functional theory (DFT), B3LYP [40], and the 3-21G* basis set (Gaussian 03W software41). The EP maps were calculated onto the molecular surface of compounds using GaussView 05 software.42 The interpretation of EP maps is based on a color scheme, where regions having higher electronic density distribution are presented as intense red color (negatively charged) whereas regions with lower electronic density distribution are shown as intense blue color (positively charged). Since the EP property has been calculated onto the molecular surface of each compound, their molecular shapes were also assessed. Moreover, the molecular volume (intrinsic molecular property) of each compound considering the van der Waals radii was also calculated employing Discovery Studio Visualizer 4.0 software.43 Of note, molecular shape and electronic properties are among the primary molecular properties in the ligand-receptor recognition process.
The authors thank Dr. Chudzinski-Tavassi A.M. for allowing the use of the CLC Drug Discovery Workbench 2.4 license installed on the workstation at the Biochemistry and Biochemistry Laboratory, Butantan Institute, São Paulo, SP, Brazil. Da Silva, B.A.V.G. and Kodama, R.T. also thank BNDES (#13.2.0711.1/2013) and São Paulo Research Foundation (FAPESP, process #15/13124-5), respectively, for their scholarships.
The authors declare no conflict of interest.

©2018 Pasqualoto, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.