eISSN: 2379-6367


Research Article Volume 9 Issue 3
1Department of Industrial and Physical Pharmacy, College of Pharmacy, Purdue University, USA
2Abbott Laboratories, USA
3Bristol-Myers Squibb Research Institute, Discovery Pharmaceutics and Analytical Sciences, USA
4Novo Nordisk Research Center, USA
5Eli Lilly and Company, Lilly Corporate Center, USA
Correspondence: Gregory T Knipp, Ph.D., Department of Industrial and Physical Pharmacy, College of Pharmacy, Purdue University, 575 Stadium Mall Dr. West Lafayette, IN 47907-2091, Tel 765-494-3765, Fax 765-494-6545
Received: June 08, 2021 | Published: June 18, 2021
Citation: Lindley DJ, Carl SM, Mowery SA, et al. Evaluation of human peptide/histidine transporter 1 (hPHT1/SLC15A4) function: transport kinetics utilizing a HPHT1 shRNA stably transfected knockdown in the hCMEC/D3 blood-brain barrier cell line. Pharm Pharmacol Int J. 2021;9(3):109-118. DOI: 10.15406/ppij.2021.09.00335
Purpose: Investigations of the Proton-dependent Oligopeptide Transporter Superfamily have focused mainly on hPepT1 and hPepT2, with little emphasis on the Peptide/Histidine Transporter isoforms (hPHT1 and hPHT2).1 The studies presented here describe the utilization of small-hairpin RNA (shRNA) technology to establish stable hPHT1 knockdown (KD) and missense (Control) in vitro human blood-brain barrier (BBB) immortalized cell lines (hCMEC/D3).
Methods: Comparative regulation of endogenous transporter mRNA expressions in the hCMEC/D3-hPHT1- Knockdown cells versus the hCMEC/D3-control cells were examined utilizing SuperArray’s 84 drug transporter qRT-PCR array. Monolayer integrity was verified using mannitol and urea. Several POT substrates were examined for hPHT1 specificity using bi-directional permeability studies across these cell lines.
Results: The mRNA transcripts of several drug transporter genes were up or down regulated in the hCMEC/D3-hPHT1-Knockdown cells compared to the hCMEC/D3-Control. Apical→Basolateral (A→B) studies demonstrated increased permeability of valacyclovir and benzylpenacillin in the hCMEC/D3-hPHT1-Knockdown cells comparative to the hCMEC/D3- Control cells. Additionally, several POT substrates including the newly identified hPHT1 substrate, thiamine showed an increase in Basolateral→Apical (B→A) permeability.
Conclusions: These results suggest that hPHT1 may function differently than PepT1 and PepT2, particularly in the BBB. In addition, hPHT1 may play a neuroprotective role, effluxing substrates out of the basement membrane in the BBB.
Keywords: hCMEC/D3, human proton-dependent oligopeptide transporters, PCR, solute carrier transporters, thiamine, peptide/histidine transporter 1, PHT1, blood-brain barrier
ABC, ATP-binding cassette; A, apical; B, basolateral; BLQ, below the limit of quantitation; EGM, endothelial growth medium; GAPDH, glyceraldehyde-3 phosphate dehydrogenase; GI, gastrointestinal; HBSS, Hank’s balanced salt solution; hPepT, human oligopeptide transporter; hPHT, human peptide/histidine transporter; MES, 2-(N-morpholino)ethanesulfonic acid; MDR, multidrug resistance; MRP, multidrug resistance-like protein; qRT-PCR, quantitative reverse transcriptase-polymerase chain reaction; P-gp, P-glycoprotein; POT, proton-dependent oligopeptide transporter; RP L13a, ribosomal protein L13a; SLC, solute carrier transporter
It has traditionally been assumed that peptides and proteins do not cross the blood-brain barrier (BBB) due to the barrier’s inherent microvessel endothelial cells’ restrictive physiology.1,2 In the 1960s, researchers demonstrated distinct differences between the capillary bed that forms the BBB and those in other peripheral tissues; e.g., lack of pores (fenestrae), presence of tight junctions, and the lack of fluid phase pinocytosis were all identified characteristics of the BBB.2 The restrictive nature not only prevents leakage of proteins into the BBB but also prevents the passage of much smaller molecules (e.g., lanthanum, <139 Da) as well.3 Restricting the penetration of certain molecules from circulation can be advantageous to protect the brain from potentially toxic compounds into a highly susceptible neuronal environment. However, it is also critical that the BBB allows the passage of nutrients and other endogenous compounds to nourish and enable the proper functioning of the brain.4 This is accomplished through the presence of transporters on the BBB that allow the influx of nutrients including vitamins, minerals, glucose, fatty acids, and peptides. Additionally, the BBB also has significant expression of multidrug resistance-conferring efflux transporters (e.g. P-gp, BCRP, MRP2) to exclude more lipophilic, potentially harmful compounds.5,6 Although not highlighted here, the BBB also presents a significant metabolic barrier to protect against xenobiotic permeation into the brain as well, a function that may also be enhanced by surrounding astrocytes and pericytes. Taken together, the BBB represents one of the most complex systems in physiology, designed to protect and nourish the brain.
It was demonstrated in 1979 that secretin injected into the brain stimulated pancreatic secretions into the gut at a fraction of its effective dose demonstrating that large peptides would have to cross the BBB to elicit its therapeutic effect.7 It is currently known that secretin among many other oligopeptide/protein-based compounds such as leptin, insulin, ghrelin, amylin, δ-sleep-inducing peptide, enkaphalin analogs, and pancreatic polypeptide must all cross the BBB to exert their effect on their receptors within the brain.7 In fact, many peptides in excess of 600kDa in molecular weight are transported across the BBB, although many of these larger oligopeptides and proteins are trafficked via receptor-mediated endocytosis. (5) Not only are hormonal or other regulatory molecules such as peptides and proteins actively transported into the BBB, but this process can also occur at specific locations within the brain.8,9 One mechanism used to exploit BBB large molecule transporters to enhance brain exposure to drugs is the “Trojan Horse” strategy.10 This involves coupling a compound that may not readily access the brain to a compound designed for easy entry via either transporter or a receptor-mediated endocytotic pathway. The success of BBB drug delivery approaches, such as those described above, rests on understanding the specific affinities/capacities of various substrates for these transporters and receptors expressed on the microvessel endothelial cell apical surface. Furthermore, these compounds may include single amino acid, di and tripeptide-based analogs where specific transport across the BBB via several different pathways potentially including PHT1, as described in this manuscript.
Current in vitro primary cell models used to evaluate the permeation of substrates across the blood-brain barrier can be labor-intensive to isolate, possess a high paracellular permeation that can obfuscate transcellular pathway delineation, and/or may not be representative of the human BBB.11-13 One particular study noted the variability in expression and transport kinetics associated with P-gp in various BBB models: MDCK-MDRI, primary cultured bovine brain microvessel endothelial cells (BBMEC) and freshly isolated bovine brain microvessels (BBM).14 More recently, a novel immortalized BBB cell line, the hCMEC/D3 human primary cell line, has been developed and characterized.15,16 The hCMEC/D3 cell line was developed by a lentivirus- mediated co-transfection of hTERT (human telomerase catalytic unit) and the SV40 T antigen of primary isolated human brain endothelial cells followed by subcloning and clonal characterization. One significant advantage of this cell line is that it retains human BBB endothelial cell characteristics, even in the absence of glial cell co-culturing.15 It was previously noted by our laboratory that although efflux transport had been characterized with respect to this cell line, influx transporters had not been investigated to our knowledge.1 Specifically, we were interested in characterizing the basal expression and function of proton-dependent oligopeptide transporter Superfamily isoforms in the hCMEC/D3 model.17 The results indicated that little to no PepT1 (SLC15A1) and PepT2 (SLC15A2) mRNA expression was observed in the hCMEC/D3 cells which corresponded to the human BBB, whereas PHT1 (SLC15A4) and PHT2 (SLC15A3) were expressed at relatively high levels.18-20 To that end, it was concluded that the hCMEC/D3 model may be a nice surrogate to study PHT1 and PHT2 mediated peptide and peptide-based drug transport across the BBB.
The studies described in this manuscript focus on the utilization of the hCMEC/D3 cell line to transfect and stably knockdown (KD) hPHT1 expression for functional evaluation. The utilization of shRNA has become a standard tool to study gene function21-23 and nicely compliments over-expression transfected cell lines for evaluating function (unpublished data). The initial functional investigation of PHT1 was performed with the rat isoform mRNA injected into Xenopus Oocytes, where it was demonstrated to have an affinity and capacity for di- and tri- peptides and the single amino acid histidine.20 Similarly, transiently transfected, non-polarizable COS-7 cells demonstrated a pH-dependent effect where hPHT1 was involved in the transport of various peptides and peptide-like compounds such as valacyclovir, Gly-Sar, carnosine, and histidine, similar to what was reported previously.24 Conversely, the transiently transfected data was not supported by research in stably transfected COS-7 cells,19 which led us to hypothesize that selective pressure (antibiotic incorporation) in this system may be a confounding factor. Although PepT1 has been previously demonstrated to be characterized this way in MDCKII cells,25,26 we hypothesized that hPHT1 characterization was gene (human splice variants may be required) and cell line dependent, and as a result, not as simple to determine. It has been established that an hPepT1 splice variant can alter its pH-sensitivity, thus lending support to our hypothesis.27-29 Thus, here we present results utilizing hPHT1 shRNA transfection and KD for characterization of its function. The hCMEC/D3 cells were selected for characterization based upon the cell line’s human origin and its relatively high level of hPHT1 mRNA expression. The results of the studies outlined here suggest hPHT1 plays an important role in basolateral efflux of peptides and peptide-based substrates. Furthermore, hPHT1 KD unexpectedly affected mRNA expression of a number of other pharmaceutically relevant transporters.
Materials
Endothelial Growth Medium-2 (EGM-2) and supplements were obtained from Lonza (Walkersville, MD). The West Femto SuperSignal Detection kit and the BCA protein quantitation reagents were obtained from Pierce (Thermo Electron, Rockford, IL). The qRT-PCR drug transporter arrays (Catalog # PAHS-070) and RT2 First-Strand kit (C-03) were obtained from SA Biosciences (Frederick, MD). The Absolutely RNA® isolation kits, Brilliant II SYBR Green QRT- PCR 1-Step Master Mix and AffinityScript® qRT-PCR cDNA synthesis kit were obtained from Stratagene (La Jolla, CA). Radiolabeled compounds were obtained from Moravek Biochemicals (Brea, CA). The MISSION® shRNA transduction particles, control transduction particles, glycerol stocks, penicillin-streptomycin solution, trypsin, DNA purification kits, and Phosphate Buffered Saline (PBS), and all other chemicals and reagents were obtained from Sigma Chemical Company (St. Louis, MO) unless otherwise noted.
Cell culture
The hCMEC/D3 cell line was kindly donated by Dr. P. Couraud from the Institut Cochin Université Paris, CNRS Descartes, Paris, France. The hCMEC/D3 cell line was cultured as described previously.1,15 Briefly, the cells were maintained at 37°C, 5% CO2, and 90% RH in a modification of the proprietary EGM-2 from Lonza (Walkersville, MD), containing 2.5% fetal bovine serum, penicillin, streptomycin, 0.1% fibroblast growth factor (FGF), 0.01% hydrocortisone and 0.025% each of insulin-like growth factor, vascular endothelial growth factor (VEGF), and endothelial growth factor (EGF). Cells were passaged into collagen coated culture flasks every 3-4 days at approximately 80%-90% confluence.
Stable transfection of hPHT1 shRNA
Five individual clones of hPHT1 small-hairpin RNA (shRNA) sequences were designed and obtained from Sigma’s MISSION® shRNA gene silencing laboratory (Table 1). The hPHT1 shRNA sequences were inserted into apLKO.1-puro plasmid that contains a puromycin resistance gene for stable selection. In addition, a control plasmid was obtained that contained a missense or “scrambled” sequence that was verified by Sigma to not interfere with other mammalian genes. The control and the five separate shRNA sequences were separately transduced into the hCMEC/D3 cells. The cells were established and the optimal hPHT1 shRNA transfect (shRNA sequence #2) was selected based on RNA and protein level knockdown efficiency when contrasted with the control expression levels (data not shown). Therefore, hPHT1 (Sequence #2) shRNA lentiviral vector was stably transfected into the hCMEC/D3 cells.
Name or Identification Number |
Sequence 5’-3’ |
1- TRCN0000044748 |
CCGGCGGCTGCTATTTGAACTATTACTCGAGT |
AATAGTTCAAATAGCAGCCTTTTTG |
|
2- TRCN0000044750 |
CCGGGCAGTCACACAGACTTTGGTACTCGAGT |
ACCAAAGTCTGTGTGACTGCTTTTTG |
|
3- TRCN0000044751 |
CCGGCGTGGGCATGTTCTTTGTCATCTCGAGA |
TGACAAAGAACATGCCCACGTTTTTG |
|
4- TRCN0000044752 |
CCGGTGTTCAAGATACTGACGTATTCTCGAGA |
ATACGTCAGTATCTTGAACATTTTTG |
|
5- TRCN0000044749 |
CCGGGCATTGGAGTCTTTCAGCAATCTCGAGA |
|
TTGCTGAAAGACTCCAATGCTTTTTG |
Table 1 PHT1 MISSION® shRNA individual clone sequences inserted into the MISSION® pLKO.1-puro vector obtained from Sigma Chemical Company
The hCMEC/D3 cells were seeded at a density of 2.0 x 105 cells/cm2 into collagen coated 24 well plates. At day one post seeding, the cells were transfected in triplicate in fresh medium with 8µg/mL hexadimethrine bromide and lentiviral particles containing the MISSION® pLKO.1-puro/PHT1 vector (Sigma, St. Louis, MO) at 10,000 TU/well. In addition, lentiviral particles containing the MISSION® pLKO.1-puro vector with a missense, scrambled shRNA sequence with no identity to known mammalian mRNA sequences were transfected at 10,000 TU/well as a negative control (Control). The medium was changed the day following transfection and replaced with fresh medium. On day four, 8µg/mL of puromycin was added to the media based upon the results of a concentration-dependent puromycin cytoxicity study conducted to determine the minimum lethal concentration for the endogenous cells (data not shown). This concentration enabled selection of only the cells expressing the transfected vector. The hCMEC/D3-hPHT1 knockdown (abbreviated KD from here on) and hCMEC/D3-Control cells were passaged every 3-4 days in media containing 8µg/mL puromycin throughout our studies to maintain selective pressure (based on a puromycin concentration dependent cytotoxicity curve, data not shown).
Evaluation of gene knockdown using real-time PCR
Briefly, hCMEC-hPHT1 shRNA KD and hCMEC/D3-Control cells (hCMEC/D3-hPHT1 KD and hCMEC/D3-Control cells, respectively) were seeded in collagen coated 6 well plates in triplicate at a density of 2.0 x 105 cells/cm2 and maintained in 8µg/mL puromycin. The media was changed on day 1 post-seeding and replaced with fresh medium containing either 0, 4 or 8µg/mL puromycin. mRNA was harvested the following day (day 2 post-seeding). Total RNA was isolated using Absolutely RNA® isolation kits per the manufacturer’s protocol and as described previously by our laboratory.30 Upon isolation, the total RNA was quantified and the relative purity was assessed using gel electrophoresis separation and densitometric evaluation of the 28S and 18S bands. In addition, reaction conditions and primer concentrations (Table 2.1) for qRT-PCR were optimized using an MX3000P QPCR system from Stratagene (La Jolla, CA). Complimentary DNAs (cDNAs) were generated using AffinityScript® from Stratagene (La Jolla, CA). Quantitative Real-Time PCR (qRT-PCR) reactions were performed with the Brilliant II SYBR Green Master Mix (Stratagene) and dissociation curves of the final products were obtained to check amplification purity. The Ct values for PHT1 obtained from the amplification plots were standardized to the housekeeping gene GAPDH as an endogenous control. qRT-PCR theory and data analysis have been described previously in our laboratory.31
Substrate |
pH Gradient |
Apparent A-B Permeability, cm/sec Coefficient (A-B Mannitol Corrected) |
Apparent B-A Permeability Coefficient, cm/sec (B-A Mannitol Corrected) |
|||
Donor |
Receiver |
Control (SD) |
Knockdown (SD) |
Control (SD) |
Knockdown (SD) |
|
Valacyclovir |
Iso pH 7.4 |
0.764 (0.03) |
0.870 (.04)* |
0.728 (0.02) |
0.929 (0.07)** |
|
5.5 |
7.4 |
0.709 (0.02) |
0.740 (0.01) |
0.593 (0.02) |
0.720 (0.07)* |
|
7.4 |
5.5 |
0.745 (0.03) |
0.787 (0.03) |
0.675 (0.01) |
0.727 (0.03)* |
|
8.5 |
7.4 |
0.787 (0.03) |
0.828 (0.01) |
0.683 (0.07) |
0.900 (0.02)** |
|
7.4 |
8.5 |
1.06 (0.07) |
0.96 (0.07) |
1.070 (0.20) |
0.831 (0.12) |
|
Table 2 Mannitol corrected A-B and B-A permeability coefficients for valacyclovir transport in the hCMEC/D3-hPHT1 knockdowns and hCMEC/D3-Control cells at varying pH gradients
Results are presented as mean±SD of 3 replicates. * p<0.05, ** p<0.01; *** p<0.001 as determined by two-tailed unpaired students t-test.
Total protein isolation
Total protein was isolated from hCMEC/D3-hPHT1 KD and hCMEC/D3-Control cells cultured under varying concentrations of puromycin (0, 4 or 8µg/mL) using a modified radio-immunoprecipitation assay (RIPA) buffer. Briefly, cells were lysed in RIPA buffer containing 0.1% Triton-X and 1µg/mL protease inhibitor cocktail, as described previously by our laboratory.26 Final protein concentration was determined by bicinchoninic acid (BCA) assay according to the manufacturer’s protocol (Pierce Biotech).
Western blotting
Forty µg of the total protein cell lysate from each of the hCMEC/D3-hPHT1 KD and hCMEC/D3-Control cells were electrophoretically separated on 10% SDS polyacrylamide gels, as described previously by our laboratory.26 Briefly, the gel separated cellular proteins were then electrophoretically blotted onto PVDF membranes. The membranes were blocked with 5% powdered nonfat dry milk (NFDM) in TBST (1X tris-buffered saline (TBS) with 0.1% Tween20) overnight at 4°C. The blots were then probed with a 1:200 dilution of a custom designed hPHT1 rabbit polyclonal antibody (Invitrogen, Inc.) in 5% NFDM in TBST for 1hr at room temperature. Following primary antibody incubation, the blots were washed 3 separate times (10 min per wash) using TBST. The blots were then probed with a horse-radish peroxidase (HRP) -conjugated secondary anti-rabbit monoclonal antibody (1:5000 dilution) in 2% NFDM in TBST for 1hr at room temperature, washed three additional times (10min per wash), and then incubated with the West Femto Supersignal Chemiluminescent Reagents (Pierce Chemical Co.) for 1min. The resultant chemiluminescent immunoreactions were visualized using a Biorad ChemiDoc XRS chemiluminescent imaging system equipped with a 12-bit Peltier cooled digital CCD camera. Data captured by the camera were analyzed using the Biorad QuantityOne Software Suite. Blots were stripped, re-probed with a 1:5000 dilution of the biological/loading control GAPDH, and detected in the same manner described above.
Permeability studies
All transport studies were conducted in triplicate for 90 minutes using either the stably transfected hCMEC/D3-hPHT1 KD or hCMEC/D3-Control cell lines, as described previously by our laboratory.25,26 Transport studies were performed in both the apical to basolateral (A→B) and basolateral to apical (B→A) directions. Studies were performed across 12mm tissue culture treated, collagen-coated polyester membranes (0.4μm pore size, 3460 Transwells®). Cells were seeded at 2 x 105 cells/cm2 and grown for 2 days prior to the transport study, according to the previously published protocol.1,15 The puromycin containing growth medium was replaced with fresh media with no puromycin the following the first day of seeding. Prior to the transport study, the culture medium was removed and the cells were washed twice with the appropriate pre-warmed transport buffer, and then equilibrated in transport buffer for 15 minutes. A stock transport buffer solution containing the substrate of interest was added to the donor side of the cells for each respective study and the appearance kinetics of the respective compounds were determined in the receiver side. Cells were maintained on a rocker platform (30 rpm) at 37°C throughout the study to disrupt unstirred boundary layers. [14C] Mannitol and [14C] urea transport kinetics were used to ensure cell monolayer integrity prior to performing POT substrate transport studies. The appearance of the selected POT substrates was determined in the receiver side at 15, 30, 45, 60 and 90 minutes. Apparent permeability coefficients have been determined using equation 1:
Papp = (dq/dt)*(1/ACo) (1)
Where dQ/dt is the steady state appearance rate in the receiver compartment, C0 is the initial concentration in the donor compartment, and A is the surface area of exposed membrane (cm2). Sink conditions were maintained throughout each study.
Valacyclovir pH gradient studies
Bidirectional permeability studies were performed as described above with the exception of the buffer possessing several different pHs in the donor and receiver chambers to establish the pH gradients. The hCMEC/D3-hPHT1 KD and hCMEC/D3-Control cells were seeded as described above and washed twice with corresponding donor and receiver buffers (MES Ringer’s pH 5.5, HBSS pH 7.4 or Tricine Ringer’s pH 8.5) followed by a pre-incubation period of 15 minutes in the corresponding transport buffers to establish the pH gradient. Cells were then incubated with 1µCi/mL [3H]valacyclovir added to the corresponding donor chamber. Both A→B and B→A studies were performed under the following pH gradients: Donor pH 5.5, Receiver pH 7.4; Donor pH 7.4, Receiver pH 5.5; Donor pH 8.5 Receiver pH 7.4; and Donor pH 7.4, Receiver pH 8.5.
qRT-PCR drug transporter arrays
Total RNA was extracted per the manufacturer’s instructions as described above in the PCR methods section. Transporter qRT-PCR arrays (SA Biosciences®) profiling the expression of 84 SLC and ABC transporter member sequences that are relevant to drug ADME were performed with the hCMEC/D3-hPHT1 KD and hCMEC/D3-Control cells to assess if there were changes in drug transporter expression with respect to the knockdown of hPHT1, as described by our laboratory previously.1,30,31 The cDNA’s were prepared utilizing the SA Biosciences RT2 kit (C- 03) according to the manufacturer’s protocol. All qRT-PCR arrays were performed in triplicate, as described previously by our laboratory.1,30,31 QRT-PCR was performed using the Brilliant II SYBR Green Mastermix from Stratagene. Results were normalized to the average of five housekeeping genes; β-actin, β-2 microglobulin, glyceraldehyde-3 phosphate dehydrogenase (GAPDH), hypoxanthine phosphoribosyltransferase 1, and ribosomal protein L13a. The limit of quantitation was set at a Ct≥35 and any genes expressed in this range were excluded and set as below the limit of quantitation (BLQ).
Thiamine inhibition
Inhibition of [3H]valacyclovir B→A transport in the presence or absence of 1mM thiamine was evaluated in the hCMEC/D3-hPHT1 KD and hCMEC/D3-Control cells utilizing the same permeability methodology as described above. Briefly, cells were washed twice on the day of the study and equilibrated for 15 minutes in transport buffer (with or without 1mM thiamine). Buffer containing [3H]valacyclovir (1μCi/mL), was then added to the respective chamber to assess B→A transport in the presence, or absence of inhibitor. All studies were performed in triplicate and results are reported as mean ± standard deviation.
Statistical analysis
All results were statistically analyzed using an unpaired, two-tailed student’s t-test at a 95% confidence level to determine potential differences between data sets. Statistical significance was denoted in the figures using the following notations: * for p≤0.05, ** for p≤0.01, and *** for p≤0.001.
hPHT1 knockdown efficiency utilizing qRT-PCR
In preliminary studies, we evaluated the effect of various levels of puromycin concentration on hPHT1 expression if it was removed the day before a respective study would be conducted (i.e. expression analysis or permeability study) and determined that the optimum selective pressure to maintain hPHT1 KD in media was at 8µg/mL puromycin (data not shown). In this study, the cells were switched from the selective pressure media containing 8µg/mL of puromycin to media containing 0, 4 or 8µg/mL puromycin 24 hours prior to RNA and protein isolation (extraction on day 2 post-seeding). Allowing the cells to equilibrate for 24 hours without puromycin would be ideal since this compound inhibits protein synthesis. Additionally, puromycin has a structure similar to some of the compounds (e.g. Valacyclovir) that were going to be evaluated in the study. It was therefore prudent to exclude this as a potential confounder by allowing sufficient equilibration time. Figure 1(A) illustrates hPHT1 mRNA knockdown efficiency in these cells subjected to varying puromycin concentrations utilizing qRT-PCR, at an initial passage. These results demonstrate that hPHT1 mRNA was significantly down-regulated in the hCMEC/D3-hPHT1 KD cells compared to the hCMEC/D3-Control cells at the different puromycin concentrations. Furthermore, there no statistically significant differences were observed in hPHT1 mRNA expression at the 0, 4, and 8µg/mL puromycin concentrations. In addition, utilizing the ΔΔCt method of qRT-PCR analysis, the hCMEC/D3-hPHT1 KD cells were compared directly to the hCMEC/D3-Control cells to demonstrate the efficiency as a percent of the control. Figure 1 (B) shows an approximate 80% reduction in mRNA transcripts in the hCMEC/D3-hPHT1 KD (8µg/mL puromycin concentration) was observed comparative to the hCMEC/D3-Control cells at a passage prior to experimentation. Finally, hPHT1 mRNA expression was examined in the hCMEC/D3-hPHT1 KD and hCMEC/D3-control cells that were cultured until the end of our experiments (approximately 10 passages) and were determined to have nearly identical knockdown efficiencies at each concentration and time point as those that were observed in the initial passage measurement (~80%, data not shown).
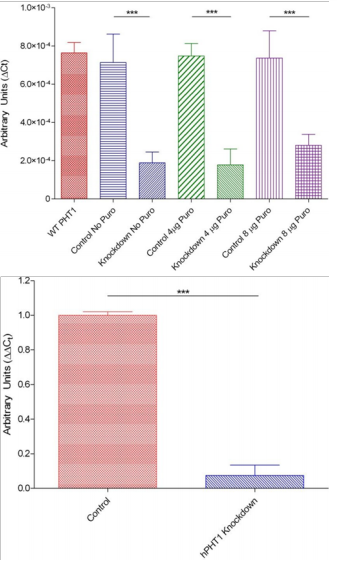
Figure 1 (A) Illustration of hPHT1 mRNA expression by qRT-PCR in the hCMEC/D3-Control and hCMEC/D3-hPHT1 knockdown cells presented as ΔCt’s using GAPDH as the housekeepeing gene and (B) Illustration of hPHT1 mRNA knockdown efficiency (extrapolated from Figure 1(A)) qRT-PCR in the hCMEC/D3-Control (8µg/mL puromycin) and hCMEC/D3-hPHT1 Knockdown (8µg/mL puromycin) cells presented as a ΔΔCt using the hCMEC/D3-Control cells as a calibrator and GAPDH for housekeeping. Cells were seeded in triplicate at a density of 2 x 105 cells/cm2 on day 1. On day 2 the media was changed and replaced with fresh media containing 0, 4, or 8µg/mL puromycin. RNA was extracted and hPHT1 expression was evaluated on day 3. Wild type hCMEC/D3 hPHT1 expression is included for comparison. Figures represent hPHT1 mRNA expression at an initial passage before initiation of experimentation. ***-p≤0.001.
Western Blot Analysis on the protein lysates isolated from the hCMEC/D3-hPHT1 KD and hCMEC/D3-Control cells cultured with different puromycin concentrations was utilized to evaluate if hPHT1 protein expression knockdown efficiencies were analogous to the observed mRNA changes. Figure 2 (A) illustrates the Western Blots of the hCMEC/D3-Control (C) and hCMEC/D3-hPHT1 KD (K) at the various puromycin concentrations indicated. These results demonstrate an apparent decrease in hPHT1 protein expression in the knockdowns compared to controls (at each puromycin concentration indicated) at the initial passage prior to conducting our transport experiments. The blots were stripped and re-probed with GAPDH to ensure equal loading of the lysates. Protein expression was evaluated at the final passage-post experimentation (Figure 2 (B)), which illustrated similar results.
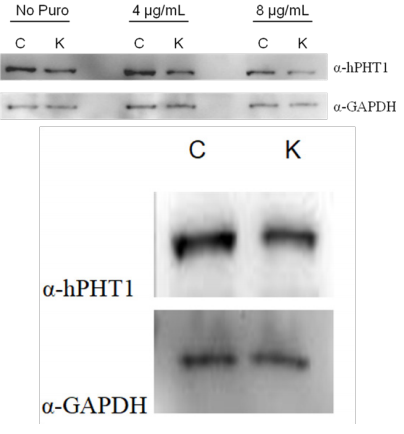
Figure 2 Western Blotting results illustrating hPHT1 protein knockdown in the hCMEC/D3- hPHT1 knockdown cells (K) compared to the hCMEC/D3-Control cells (C) utilizing a polyclonal custom-designed antibody from Invitrogen at (A) an initial passage before initiation of experimentation and (B) a final passage post-experimentation. The blots were stripped and re- probed with GAPDH as a biological and loading control. Cells were seeded at a density of 2 x 105 cells/cm2 on day 1. On day 2 the media was changed and replaced with fresh media with different puromycin concentrations shown above. Protein lysates were obtained on day 3.
Permeability studies
To assess the effect of transduction/hPHT1 knockdown on the cell monolayer integrity, we evaluated the permeability of the small, neutral paracellular markers mannitol and urea. The apparent permeability coefficients were not significantly different between the A→B and B→A directions in either the KD or Control cell lines for each substrate (data not shown). Although not statistically significant, accounting for the subtle differences in the paracellular permeation of mannitol and urea can be important. Therefore, permeability studies conducted with hPHT1 substrates were normalized to mannitol as a control.
After confirming the monolayer integrity was not altered, the transcellular permeation of selected POT substrates in both the A→B and B→A directions were normalized to the respective A→B or B→A mannitol value to account for directional differences in paracellular flux. Figure 3 (A) and (B) illustrates respectively A→B and B→A permeation of various POT and other potential hPHT1 substrates through the hCMEC/D3-hPHT1 KD and hCMEC/D3-Control cells. There was an apparent lack of statistically significant differences between the two cell lines in the A→B mannitol corrected permeability of histidine, carnosine, glycylsarcosine (gly-sar), 5- aminolevulinic acid (ALA), valproic acid (VPA), and γ-aminobutyric acid (GABA), as illustrated results in Figure 3 (A). In contrast, statistically significant differences were observed for the mannitol corrected permeability of valacyclovir and benzylpenicillin, with the hCMEC/D3-hPHT1 KD having higher A→B permeation in contrast to the hCMEC/D3-Control cells. The results in Figure 3, the B→A direction, illustrate no statistically significant differences between the two cell lines for mannitol corrected carnosine, VPA, GABA and benzylpenicillin permeation. In contrast, statistically significant differences were observed revealing higher mannitol corrected permeability coefficients for histidine, valacyclovir, gly-sar, and ALA being observed in the hCMEC/D3-hPHT1 KD compared to the hCMEC/D3-Control cells. The intracellular accumulation of these substrates was also measured in the hCMEC/D3-hPHT1 KD and hCMEC/D3-Control cell lines to establish mass balance and potentially as a means to evaluate hPHT1 substrate specificity. No appreciable intracellular accumulation was observed in either direction in both cell lines for most of the substrates with the exception of histidine, which was approximately 2 fold lower in the hCMEC/D3-hPHT1 KD compared to hCMEC/D3-hPHT1-control cells.
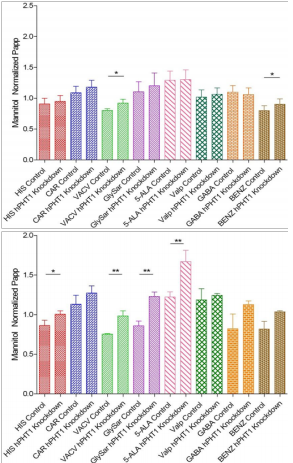
Figure 3 Mannitol-corrected apparent permeability coefficients for various substrates across either hCMEC/D3-Control or hCMEC/D3-hPHT1 knockdown cells in (A) the A→B direction and (B) the B→A direction. Results in the A→B direction indicate no discernable differences between the two cell lines for all substrates except for valacyclovir and benzylpenicillin with transport being higher in the hCMEC/D3-hPHT1 knockdown cells. The B→A data illustrates differences in apparent permeability for histidine, valacyclovir, Gly-Sar and 5-aminolevulinic acid in the hCMEC/D3-hPHT1 knockdown cells compared to the hCMEC/D3-Control cells. Results are presented as mean±SD of 3 replicates. * p<0.05, ** p<0.01; *** p<0.001 as determined by two-tailed unpaired students t-test.
Valacyclovir permeability with various pH gradients
To investigate if a proton-dependent gradient plays a role in hPHT1 mediated translocation of substrates across the membrane, permeability studies were performed with valacyclovir utilizing selected extracellular pH gradients in both the hCMEC/D3-hPHT1 KD and hCMEC/D3-control cells. The results of those studies are described in Table 2. Statistically significant differences were not observed in the A→B direction under the various pH gradients, with the exception of the iso pH of 7.4. Interestingly, statistically significant differences were identified in valacyclovir transport in the B→A direction with higher transport observed in the hCMEC/D3-hPHT1 KD compared to the hCMEC/D3-Control cells. Specifically transport was higher in the knockdowns at iso pH 7.4; B(pH 5.5)→A(pH 7.4); B(pH 7.4)→A(pH 5.5) and B(pH 8.5)→A(pH 7.4). Since the cytosolic pH is approximately pH 7.4, pH gradients are also established as a driving force specifically for those conditions where the apical or basolateral pH are higher than pH 7.4.
Drug transporter expression using qRT-PCR arrays
qRT-PCR arrays containing 84 pharmaceutically relevant transporters were obtained from SA biosciences, and were utilized to evaluate the expression of these transporters in the hCMEC/D3-hPHT1 KD and hCMEC/D3-Control cells, as described previously by our laboratory.1,30 These studies were conducted to provide an indication of the regulation of mRNA expression for other transporters as a potential compensatory response to hPHT1 knockdown. Selective pressure was removed in this experiment the day before harvesting to remain consistent with the procedure used for the permeability studies (see above). Figure 4 illustrates those mRNA transcripts that were significantly up- or down-regulated in response to hPHT1 knockdown. As observed in Figure 4, transporter mRNAs that demonstrated expressional changes in response to the hPHT1 down-regulation when comparing the hCMEC/D3-KD to the hCMEC/D3-Control cells included the up-regulation of hPHT2 (SLC15A3), the Breast Cancer Resistance Protein (BCRP; ABCG2), the neutral and basic amino acid transporter (SLC3A1), and the apically expressed thiamine transporter (THTR2, SLC19A3). In addition, MRP1 (ABCC1), MRP3 (ABCC3), and Cu2+ transporting polypeptide (ATP7A) were also down-regulated.
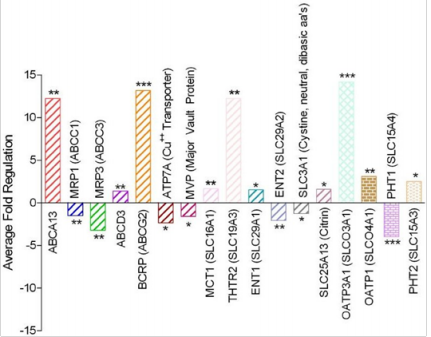
Figure 4 Average fold regulation of drug transporter genes utilizing qRT-PCR arrays in the hCMEC/D3-hPHT1 Knockdown (No puromycin addition on Day 2) cells using the hCMEC/D3-Control (No puromycin addition on Day 2) cells as the calibrator (Ct). Data was selected based on those genes that were statistically significantly up/down regulated. * p<0.05, ** p<0.01, and *** p<0.001.
Thiamine inhibition and transport
As mentioned above, there were several mRNA expressional changes in drug related transporters that were associated with the hPHT1 knockdown. While the corresponding protein changes requires further validation, of particular interest to our lab was the investigation of the potential effects of the up-regulation of THTR2. Due to its structural similarity to histidine, we were interested in determining if thiamine (Vitamin B1) was a potential hPHT1 substrate. Figure 5 (A) illustrates mannitol normalized B→A valacyclovir transport in the presence and absence of 1mM thiamine in the hCMEC/D3-hPHT1 KD and hCMEC/D3-Control cells. Interestingly, a statistically significant difference in the mannitol normalized B→A valacyclovir transport in the hCMEC/D3-hPHT1 KD cells was observed when 1 mM thiamine was added and the data was contrasted with the control cells. To further investigate if thiamine possesses affinity and capacity as an hPHT1 substrate, [3H] thiamine was used in transport studies with these cells. Figure 5 (B) illustrates that thiamine may be an hPHT1 substrate since the mannitol normalized B→A permeability of thiamine is greater in the hCMEC/D3-hPHT1 KD versus the hCMEC/D3-Control cells. Consistent with previous findings, statistically significant differences were not observed in the A→B direction transport data.

Figure 5 (A) Thiamine inhibition of A→B and B→A valacyclovir transport by 1 mM nonradiolabelled thiamine. Inhibitor was not introduced to cells until after a 15 minute equilibration period (buffer + inhibitor). and (B) Mannitol corrected A→B and B→A permeability coefficients for [3H]Thiamine in the hCMEC/D3-hPHT1 knockdown cells compared to the hCMEC/D3-Control cells. Results are reported as mean±SD of 3 replicates. * p<0.05; ** p<0.01; *** p<0.001 using two-tailed unpaired student’s t-test.
The utilization of siRNA and shRNA mediated knockdowns to study mRNA and protein expression of isoforms of interest has gained continued interest over the last several years.21-23 For example, research performed on specific transporter isoforms using si and shRNA has proven to be a very useful tool for further identification of additional specific substrates. Although transfection systems that result in an over-expression of a protein of interest have also been widely used, they can be limited by proper post-translational modification, species differences in cell origin, and the expression levels of other endogenous cellular regulatory proteins (e.g., cellular trafficking and the presence of other transporters to meet energy requirements) to properly demonstrate function. Another advantage of this technology is that one is not relying on a gene transcription promoter from a non-human source, e.g. the cytomegalovirus (CMV) promoter, to drive mRNA synthesis, which may have helped create some challenges for the characterization of hPHT1 function.32-34 Therefore, hPHT1 shRNA mediated disruption was utilized as an alternative means to reduce protein expression in the human hCMEC/D3 BBB cell model to minimize the aforementioned potential confounding variables that may be present in over-expressed systems. This cell line provided the highest endogenous hPHT1 expression comparative to other cells lines that were examined such as Caco-2, HT-29 and MDCKII cells and has low to negligible expression of other, potentially obfuscating, peptide transporters (hPepT1 and hPepT2) that exhibit overlapping substrate specificity with hPHT1.1
The chosen hPHT1 shRNA vector transfected into the hCMEC/D3 cell line provided greater than 80% knockdown of the endogenous hPHT1 mRNA transcripts compared to the hCMEC/D3-Control cells. Additional data demonstrated that hPHT1 knockdown at the genomic was maintained from initial passaging (Figure 1A and 1B) through the final passaging post- experimentation. The reduction in hPHT1 expression was confirmed by Western Blotting in the hCMEC/D3-hPHT1 KD cells, which demonstrated lower protein levels in contrast to the hCMEC/D3-Control cells at varying puromycin concentrations in the initial passage (Figure 2A). Furthermore, similar results were observed at the final passage post-experimentation (Figure 2B). These results suggest that the hPHT1 protein knockdown was achieved and the expression differences were consistent throughout the time-course of our experiments.
The ability to delineate specific transport pathways (e.g., paracellular vs. transcellular) is highly dependent on tight junctional complex formation and pathways that lead to the subsequent polarization of the cells that result in differential transporter (based on mRNA level) expressions on the apical and basolateral membranes. Thus, it is critical to identify the effect of transfection on monolayer integrity. Mannitol and urea permeation through both the hCMEC/D3-hPHT1 KD and hCMEC/D3-Control cells were not statistically significant in either the A→B or B→A directions, suggesting that the monolayer remained similar post-transfection. Subsequent studies with the transcellular substrates were begun to identify potential hPHT1 specificity and normalized to mannitol permeation to insure that the substrates also did not alter the monolayer integrity. The substrates histidine, carnosine, Gly-Sar, benzylpenicillin, and valacyclovir were chosen either based on their POT specificity35 or data previously generated in our laboratory.24 In addition, 5-ALA (hypothesized hPepT1 and hPepT2 affinity) and VPA (purported negative hPHT1 control) were identified as potential hPHT1 substrates based on their physicochemical properties and the fact that they are known to traverse the BBB and affect neuronal function. An additional hypothesized hPHT1 substrate, γ-aminobutyric acid (GABA), was also investigated based on its structural similarity to other tested substrates and its purported role in neuronal generation.36
The results of these permeability studies were interesting when contrasted to earlier hPHT1 functional studies generated in the non-polarizable COS-7 transiently transfected cells which suggested net hPHT1 influx for various POT substrates24 and stably transfected cell lines that suggested net efflux for gly-sar.19 Figure 3A illustrates statistically significant differences in A→B transport for valacyclovir and benzylpenicillin with transport in the hCMEC/D3-hPHT1 KD cells being higher than the hCMEC/D3-Control cells. Statistically significant differences were not observed for the other compounds in the A→B direction. These results would appear to suggest a net influx function in this direction for valacyclovir and benzylpenicillin. Critical to the correct interpretation of this data is cellular localization, where these studies are ongoing in our laboratory and suggest a potential basolateral and intracellular localization.37 In addition, we cannot rule out the possibility that there is a compensatory up or down regulation of a separate transporter (e.g., hPHT2) in response to the reduction of hPHT1 expression that could explain this data.
Figure 3B illustrates the B→A transport of the same substrates across the hCMEC/D3- hPHT1 KD cells when contrasted to the hCMEC/D3-Control cells. These results illustrated higher B→A permeation in the hCMEC/D3-hPHT1 KD cells for histidine, gly-sar, valacyclovir and 5- ALA. Interestingly, we continue to identify carnosine as a non-hPHT1 substrate which is contradictory to studies performed earlier in our laboratory and with the rat PHT1 isoform by Yamashita et al.20,24 In addition, VPA, benzylpenicillin and GABA do not appear to be hPHT1 substrates in the B→A direction in these studies. Conversely, 5-ALA does appear to have affinity and capacity for hPHT1 from the results presented with the knockdowns. Collectively, these results do support a hypothesis that hPHT1 may be acting as a basolateral efflux transporter to retain substrates in the brain. These results are consistent with hPHT1 functional work conducted in our laboratory with the hPHT1 stably transfected COS-7 cells.19 It is important to note that these studies provide an indication of potential substrates for hPHT1, as well-defined substrates that have not previously been identified.
As a member of the POT family, it has been hypothesized that hPHT1-mediated transport is proton driven. To address this, positive (MES pH 5.5:HBSS pH 7.4), negative (Tricine pH 8.5:HBSS pH 7.4) and neutral (HBSS pH 7.4:HBSS pH 7.4) gradients were set up to elucidate any hPHT1 pH dependent valacyclovir transport. Table 2 presents the mannitol corrected A→B and B→A permeability coefficients for valacyclovir at the various gradients. The A→B valacyclovir permeability coefficients at the various pH gradients failed to demonstrate a statistically significant difference in permeation, except under iso pH conditions. These results suggest that hPHT1 may not be involved in apical pH dependent transport of valacyclovir. Conversely, the B→A valacyclovir permeability coefficients did illustrate higher transport in the hCMEC/D3-hPHT1 KD compared to the hCMEC/D3-Control cells. These observations further suggest that hPHT1 may be localized on the basolateral membrane and function to efflux valacyclovir into the brain (basolateral compartment). This data also implies that hPHT1 function in the hCMEC/D3 cell model may not be as dependent on extracellular pH as originally hypothesized, also consistent with the results from our COS-7/hPHT1 stably transfected cells.19 However, it is not clear if there is a mechanism through which hPHT1 may function through its own H+-driven gradient inside the cell. It is therefore paramount to understand PHT intracellular and membrane localization. It is worth noting that valacyclovir has three pKa’s of 9.43, 7.47 and 1.90 which allows it to exist in a combination of the di-cation, cationic, neutral and/or anionic forms.38 Valacyclovir would mainly exist in the cationic and neutral forms over the different pH conditions studied here, and should be considered in future hPHT1 studies with valacyclovir. Additionally, other transporters have been implicated in valacyclovir transport and their potential role here in proper data interpretation should be acknowledged.39-41 It is also important to note that cellular pH homeostasis is well maintained and its regulation is rather complex, thus complicating a clear explanation for our observations.
Our laboratory has recently utilized qRT-PCR arrays to build a more comprehensive view of transporter expression within a given cell line. These arrays were used to identify endogenous transporter expression in wild-type hCMEC/D3 cells,1 evaluate the effect of media composition on transporter expression in HT-29 cells 30 and characterize transporter/metabolism enzymes in Caco-2 cells.31 In the present study, these arrays were useful in identifying up or down regulation of transporters that may be regulated by the hPHT1 KD and serve as confounders for data interpretation by making a direct comparison between the hCMEC/D3 KD and hCMEC/D3-Control cells. The results presented here suggest that a variety of transporters’ mRNA were either up or down regulated in response to reducing hPHT1 in the knockdown cells. An interesting observation was the statistically significant up-regulation of hPHT2 (SLC15A3) in this system, potentially suggesting that hPHT2 may have overlapping, compensatory function compared to hPHT1. The presumably compensatory upregulation of hPHT2 observed here could also explain the lack of hPHT1 function observed by others.42 In addition, Cu++ transporter (ATP7A) was down- regulated suggesting that it may play a role with hPHT1.
Interestingly, the apically expressed thiamine transporter (THTR2; SLC19A3) was significantly up-regulated in response to hPHT1 knockdown in these cells. Thiamine is a water- soluble vitamin (B1) that is involved in a variety of cellular processes including the regulation of apoptosis43 and the metabolism of glucose (in the form of thiamine pryrophosphate, TPP).44 Thiamine has also been demonstrated to stabilize the membrane of newly formed neuronal cells during embryogenesis, thereby slowing developmental apoptosis.45 Others have demonstrated severe CNS effects, ranging from cell atrophy to cell death, in rats that lacked thiamine in their diet.43 The potential relationship between the knockdown of hPHT1 to the up-regulation of the THTR2 remains to be shown.
As has been suggested in this manuscript, hPHT1’s hypothesized function as a basolateral efflux transporter may render it responsible for retaining nutrients and/or neuroprotective compounds in the brain. This includes histamine, which is synthesized inside the cell via decarboxylation of histidine, and has been shown to significantly increase in the brain of thiamine-deficient rats.46,47 Taken together with the results of a recent study conducted with Pht1 knockout in mice that led to histidine accumulation in the brain, it is plausible to postulate a potentially significant role in mitigating the neuroinflammatory response.48 Furthermore, it is important to note thiamine serves as a mediator for neuroinflammation, as illustrated in vitamin B1 deficient patients with alcohol related Wernicke’s Syndrome.49 This alone would give credence to studying thiamine as a potential hPHT1 substrate, particularly when one considers the up-regulation of THTR2 in response to hPHT1 knockdown. Therefore, we decided to investigate if thiamine may inhibit valacyclovir transport in the hCMEC/D3-hPHT1 knockdowns comparative to the hCMEC/D3-Control cells in the B→A direction. These results, presented in Figure 5A, illustrated increased valacyclovir B-A permeability in the presence of 1 mM Thiamine in the hCMEC/D3- hPHT1 KD cells compared to valacyclovir B-A permeability with 1 mM Thiamine in the hCMEC/D3-Control cells, further suggesting thiamine specificity for hPHT1. This further suggests that THTR2 might be important in increasing valacylovir efflux into the brain. Additional results in Figure 5B demonstrates increased B→A thiamine transport in the hCMEC/D3-hPHT1 KD cells when contrasted to the hCMEC/D3-Control cells suggesting that it may be a substrate for hPHT1.
As mentioned above, results from our previous studies have led to confounding conclusions that may be attributable to intrinsic differences within the systems that were used to study hPHT1 function. Clearly, a cell line without high endogenous, potentially obfuscating POT isoform(s) expression, is desirable to study hPHT1 function. This was accomplished utilizing shRNA technology and hCMEC/D3 cells to knockdown endogenous hPHT1 and compare to a missense shRNA transfected control cell line. The studies presented here suggest that hPHT1 may be a basolateral efflux transporter with affinities/capacities for histidine, gly-sar, valacyclovir and 5- ALA. Furthermore, the results presented illustrate it may be necessary when performing these types of studies to identify other related gene’s that may change in response to the targeted gene that is knocked down to remove potentially confounding variables.
As discussed, thiamine transporter (THTR2) was up-regulated in response to hPHT1 KD which ultimately led to the identification of thiamine as a possible hPHT1 substrate. Further research delineating the physiological significance of thiamine’s and other related compound’s substrate affinity and capacity for hPHT1 is required to assess the significance of these findings. In addition, this further argues that it may be critically important to determine any expression/functional changes in other transporters related to the transfected gene that may obfuscate the data. Collectively, the results presented here advance the fundamental understanding of hPHT1 substrate specificity and functional relevance, although additional research is required to establish hPHT1’s role in the blood-brain barrier. The results of these findings help to substantiate the hypothesis that hPHT1 may play a crucial role in nutrient or peptide-based substrate recognition, retention, and transport across the BBB. In addition, these results may provide evidence for the rational drug and formulation design strategies for therapeutic peptide or amino acid based compounds with affinity to hPHT1 that may enhance the permeation of these agents across the BBB.
The authors would like to acknowledge Drs. Pierre Courad, BabbetteWeksler, and Ignacio Romero for providing the hCMEC/D3 cell line for these studies. The manuscript was prepared in memory of Dr. David Lindley.
The authors declare no conflicts of interest.

©2021 Lindley, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.