eISSN: 2379-6367


Research Article Volume 11 Issue 2
1Departments of Biomedical Sciences, Quillen College of Medicine, East Tennessee State University, USA
2Department of Neurology, Renmin Hospital of the Wuhan University, China
Correspondence: Dr. Meng-Yang Zhu, Department of Pharmacology, Quillen College of Medicine, East Tennessee State University, Johnson City, TN 37604, Tel (423) 439-6394, Fax (423) 439-8773
Received: June 02, 2023 | Published: June 13, 2023
Citation: Parker K, Zeng F, Zhan Y, et al. Altered DNA repair related proteins in Parkinson’s disease model VMAT2 Lo mice. Pharm Pharmacol Int J. 2023;11(2):70-85. DOI: 10.15406/ppij.2023.11.00404
DNA damage and repair processes play an important role in the pathogenesis of age-related neurodegenerative diseases such as Parkinson’s Disease (PD), as DNA repair pathways delay cell senescence and aging by maintaining genomic integrity. In the present study, the levels of DNA repair-related enzymes and proteins were examined in the brain of VMAT2 Lo mice, a PD animal model. The results demonstrated that in the frontal cortex (FC) and locus coeruleus (LC) of VMAT2 Lo mice at 2, 6, and 15 months of age, OGG1 protein levels were significantly increased. However, OGG1 protein levels in the hippocampus, substantia nigra (SN) and LC of these model mice at 18 and 23 months of age exhibited a marked reduction. This reduction of OGG1 proteins in the hippocampus and SN was accompanied by the similar diminishment of their mRNAs. Furthermore, immunochemical and immunofluorescence staining demonstrated that in most measured brain regions, the immunoreactivities of PARP1, ERCC1, XRCC1 and PCNA, four enzymes and protein related to DNA repair processes, were considerably increased in VMAT2 Lo mice at 18 and 23 months of age. These analysis results reveal the DNA oxidative damage triggers the activation of some proteins involved in the DNA repair process in this PD model, and provides important insights for the involvement of DNA repair processes in the PD development.
Keywords: DNA repair, DNA damage, oxidation stress, OGG1 dopamine, locus coeruleus, substantia nigra
BER, base excision repair; DAB, 3,3-diaminobenzidine; DA, dopamine; DAPI, 4’,6-diamidino-2-phenylindole; DSBR, double-strand break repair; ECL, enhanced chemiluminescence; ERCC1, excision repair cross 1; FBS, fetal bovine serum; FC, frontal cortex; Hip, hippocampus; LC, locus coeruleus; NER, nucleotide-excision repair; OGG1, 8-oxoguanine DNA glycosylase 1; ODs, optical densities; 8-OHdG, 8-hydro-2’dexoyguanosine; PARP1, Poly-(ADP)-ribose-polymerase 1; PBS, phosphate-buffer saline; PCNA, proliferating cell nuclear antigen; PD, parkinson’s disease; qPCR, quantitative real-time polymerase chain reaction; SDS, sodium lauryl sulfate; SN, substantia nigra; SSBs, single-strand breaks; VMAT2, vesicular monoamine transporter 2; XRCC1, X-ray cross complementing group 1
DNA damage implies any modification of DNA resulting in changes of its coding properties and functions. Higher levels of DNA damage can be the result of either increased exposure to damaging agents and/or defective repair of DNA. It is clear that compared to exposure with dangerous insults or agents, deficiency of DNA repair capacity is also an important factor contributing to pronounced neuropathology. Generally, DNA damage triggers a number of cellular responses including DNA repair, and activation of cell cycle checkpoint activity1 by which the cell cycle progression is delayed in order to facilitate DNA repair processes. Basically, the main repair processes include base excision repair (BER), nucleotide-excision repair (NER), recombinational repair, mismatch repair, and double-strand break repair (DSBR) which includes homologous recombination and non-homologous end-joining, each of which is responsible for repairing damage arising from different mechanisms.2 If damaged DNA is not properly repaired, accumulation of DNA damage can give rise to genomic instability, reduce the capacity for tissue self-renewal,3 leading to changes in gene expression, cellular dysfunction, mutations, and carcinogenesis or cell death.4,5 In particular, a decline in DNA repair capacity may be very prevalent in the central nervous system, because the brain consists largely of non-proliferative neuronal cells and is therefore particularly vulnerable to defective DNA repair that would lead to a greater steady-state level of unrepaired DNA lesions. After DNA replication and transcription are blocked, genome aberrations can occur, finally leading to the development and pathogenesis of several biological disorders including Parkinson’s disease (PD).6-8 Therefore, it is essential for cells to efficiently respond to DNA damage through coordinated and integrated DNA-damage checkpoints and repair pathways.
There is increasing evidence to suggest the neuropathogenesis in PD is related to oxidative DNA damage9 and impaired DNA repair.10 The possible role of impaired DNA repair in PD pathogenesis was indicated by an earlier study that lymphoblastoid cells obtained from sporadic PD patients were deficient in the repair of X-ray induced DNA damage.11 As mentioned above, there are several different DNA repair processes. Participating into these processes, human cells have different enzymes to counteract the damage caused by oxidative attack to nucleic acids. These include nucleoside triphosphatases that hydrolyze oxidized bases in the nucleotide pools, DNA glycosylases that remove oxidatively damaged bases from the DNA, and proteins of the mismatch repair system that remove mis-incorporated bases opposite to an oxidized base. It is striking that several such proteins have been found to be over-expressed in the substantia nigra (SN) of PD patients, pointing to an increased demand of DNA repair enzymes in the brain region primarily affected by the disease.12 For example, a significant increase in DNA damage marker proteins was shown in nigrostriatal dopaminergic neurons of PD patients paralleled by elevated levels of the DNA repair proteins such as 8-oxoguanine DNA glycosylase 1 (OGG1),13 and MUTYH glycosylase.14 PolyADP-ribose-polymerase 1 (PARP1) is another DNA repair related protein, and has been reported to be elevated in cerebrospinal fluid, brain homogenates15,16 and the serum of PD patients,17 as well as in mouse PD models.16 In addition, an increased level of X-ray cross-complementing protein 1 (XRCC1) was significantly increased in the serum of PD patients.17,18 Considering an impaired DNA repair activity may play an important role in the pathogenesis of PD, continued exploration of its characteristics in animal models is necessary.
The neural specific vesicular monoamine transporter 2 (VMAT2) is a transmembrane protein and is a key regulator of monoamine homeostasis by implication in monoamine storage, protection of neurotransmitters from oxidation and control of quantal secretion of these neurotransmitters.19 Therefore, functional failure of VMAT2 has been recognized as a contributor to the pathogenesis of PD.20 A transgenic mouse, VMAT2 Lo, presents with ~95% shutdown in endogenous VMAT2 levels.21,22 These mice are characterized by highly dysregulated transmitter homeostasis such as dopamine (DA) and norepinephrine, progressive neuronal degeneration and formation of α–synuclein containing inclusions in the SN with the ventral tegmental area unaffected.23-25 In addition, these mice exhibited non-motor features and L-DOPA-responsive motor deficits. Therefore, this model replicated essential pathogenic characteristics of PD. While many biochemical features in these model mice have been documented, little is known about the processing of the DNA damage and DNA repair ability in these animals, and no study has yet examined for DNA repair capacity in this model. It is critical to explore these pathogenic characteristics for complete establishment of a useful PD model.
Previously we have performed experiments to examine DNA damage in VMAT2 Lo mice and found that in brain regions there was a remarkable increase of DNA damage sensors such as pATM/pATR, and some DNA damage markers such as γ-H2AX and 8-hydro-2’dexoyguanosine (8-OHdG), indicating the features of DNA damage in this model. Furthermore, TUNEI assay and immunostaining confirmed the presence of apoptosis (submitted manuscript). In the present study, we examined levels of some DNA repair-related enzymes and proteins in the brain regions of VMAT2 Lo mice at different ages using biochemical and immunohistochemical assays. The results demonstrated that compared to controls, VMAT2 Lo mice showed an increase in protein levels of OGG1 in the frontal cortex (FC) and LC at the younger ages. However, model mice at 18 and 23 months of age showed a reduced level of this protein. Furthermore, immunochemical measurements revealed elevated levels of PARP1, excision repair cross (ERCC1) and XRCC1 in the related brain regions. Immunofluorescence analysis also showed an elevated levels of proliferating cell nuclear antigen (PCNA) in these brain regions. These data demonstrate that there are remarkable changes in DNA repair related enzymes and proteins in this mouse PD model, especially when age of the mouse is also considered. These alterations may lead a reduced DNA repair capacity, which may be related to the appearance of some pathological modification in this mouse model. This not only provides the evidence for the feasibility of this PD model, but also extends our understanding of this field and may have a potential effect on PD disease-modifying therapies.
Animals
The present study used both male and female VMAT2 Lo mice by courtesy of Dr. Gary Miller, Department of Environmental Health Sciences, Mailmen School of Public Health, Columbia University, New York, USA. These mice were bred in the animal facility of this university and were housed on a 12-hour light/dark cycle with food and water provided ad libitum. All animal handling procedures followed the guide approved by the Animal Care and Use Committee of East Tennessee State University (approval code: P180401), which complies with the NIH Guide for the Care and Use of Laboratory Animals.26 In the present project, different ages of mice (2, 6, 15, 18 and 23 months) and age-matched wild-type littermates were selected and were randomly assigned. A power analysis was performed to estimate the numbers of animals used in the groups by assuming the weaker of the two effects, which was also based on our preliminary studies. Although this study used both male and female mice, the sex of the animals was not separately divided in the group. Therefore, sex‐dependent effects were not assessed.
The VMAT2 Lo mice of different ages and their littermates were sacrificed by two ways based on the processing procedures. For biochemical measurements, mice were fast decapitated on the pre-determined ages without anaesthetization. Brains were removed and rapidly frozen in 2-methyl-butane on dry-ice, then stored at -80o C until dissection of the brains. For immunochemical staining, transcardiac perfusion using 4% paraformaldehyde in 0.1 mol/L phosphate buffer, pH 7.4, was performed on the mice under anesthesia with ketamine/xylazine (100mg/10mg/kg, i.p.). Then brains were collected and stored at -80oC until sectional cut was performed at 30 μm around the regions of the FC, hippocampus (Hip), SN and LC using a Leitz 1512 microtom (Leitz Co., Germany).
RNA isolation and quantitative real-time polymerase chain reaction (qPCR) analysis for mRNAs of OGG1 in brain regions
Similar methods were used for RNA isolation and qPCR analysis as described before.27 Briefly, total RNA was extracted from dissected brain regions of mice using the RNAzol reagents (Molecular Research Center, Inc., Carlsbad, CA) and converted to cDNAs by the superscript III First-Strand Synthesis Kit (Applied Biosystems/Life technologies, Forster City, CA, USA) according to the manufacturer’s protocol. The following primers were used for qPCR analysis: mouse OGG1: 5’-GAGCTGGAAACCCTACACAA-3’, and 5’-CAGGAGTCTCACACCTTGGAA-3’; mouse β-actin: 5’-CAACGAGCGGTTCCGATG-3' and 5’- GCCACAGGATTCCATACCCA-3'. The relative changes in gene expression from qPCR were analyzed as follows: first, the value of each gene of interest was normalized to β-actin (ΔCt). Then comparisons of mRNA levels of each respective gene were calculated by taking the inverse log of ΔΔCT, which resulted in the relative fold change comparative threshold cycle method (2-ΔΔCT).28 All measurements were run in triplicate, each using a separate set of cDNA aliquot.
Immunohistochemical (IHC) staining and Immunofluorescence (IF)
These methods follow similar protocols from our laboratory.29,30 All methods used the free-floating sections of different brain regions. For IHC of PARP1, ERCC1, and XRCC1, the brain sections were rinsed with PBS for 3 times, followed by blocking with 10% (v/v) nonimmune goat serum for 30 minutes. After rinsing, sections were incubated with following antibody (for PARP1, monoclonal, 1:100, ab32064, Abcam, Cambridge, MA, USA, RRID: AB_842407; for ERCC1, monoclonal, 1:250, ThermoFisher Scientific, Waltham, MA, USA, RRID: AB_1193722; for XRCC1, monoclonal, 1:100, NB500-106, ThermoFisher Scientific, Waltham, MA, USA, RRID: AB_2618029) overnight at room temperature. The next day, the sections were further exposed to biotinylated horse anti-mouse IgG. Thereafter, the sections were visualized with 3,3-diaminobenzidine (DAB) in 0.1 M Tris buffer and subsequently counterstained with hematoxylin after differentiation with hydrochloric alcohol (Vectastatin; Vector Laboratories), followed by final seal with neutral gum. The images on the slides were captured a an EVOS inverted fluorescent microscope (Advanced Microscopy Group) with attached CDD camera, and were analyzed quantitatively by ImageJ software (Rasband, US National Institutes of Health, Bethesda, http://rsbweb.nih.gov/ij, 2010).
For IF of PCNA, briefly, the sections were pre-treated in 5% bovine serum albumin in phosphate-buffer saline (PBS) supplemented with 0.4% Triton-X 100. Then these sections were incubated with primary antibody solution (mouse monoclonal antibody against PCNA, 1:500, NB500-106, RRID: AB_159246, NOVUS Biologicals, Centennial, CO, USA) overnight at 4o C. The next day, these sections were further incubated with 2nd antibodies (Alexa Fluor 488-conjugated goat anti-mouse IgG, from Abcam, Cambridge, MA, USA) after washing with 0.1 M PBS for 4 times. Then, the floating sections were mounted onto slides. Immunofluorescence labeling in the slides was observed and acquired in an Olympus microscopy (BX41 7F, Olympus Optical Co. LTD, Tokyo, Japan) with a software Spot Imaging (Version 5.3, SPOT Imaging Solution Company, Sterling Heights, MI, USA), and analyzed quantitatively by ImageJ software (Rasband, US National Institutes of Health, Bethesda, http://rsbweb.nih.gov/ij, 2010). PCNA-positive cells were also counted per millimeter square on DAB-staining images by ImageJ. The reference background levels in images of brain sections adjacent to the target region were taken as non-immunoreactive portions.
Western blot analysis
Western blotting was performed using similar methods as to that reported previously from our laboratory.29 Briefly, equal amounts of proteins (about 30 µg) from each sample were loaded into an SDS–polyacrylamide gel (10 or 15 %) and electrophoretically fractionated. Next, the proteins from the gel were electro-transferred onto polyvinylidene difluoride membranes. These membranes were incubated with the primary antibody (mouse OGG1, polyclonal, NB100-106, 1:500, NOVUS Biologicals, Centennial, CO, USA, PRID: AB_10898440) overnight at 4˚C. Then these membranes were exposed to the second antibody against rabbit or mouse. After washing, these membranes were further exposed to enhanced chemiluminescence (ECL, Amersham Life Sciences, Buckinghamshire, UK) or super ECL (Sigma Chemical Co., St Louis, MO, USA). G:Box Imaging (Fyederick, MD, USA) was used to visualize the immunoreactive bands. To verify the equal loading and transfer effectivities, an immunoreactive band of ß-actin was examined via similar incubation processes after stripping the membranes of the previous protein-antibody complexes and then using monoclonal antibody against ß-actin (1:10,000 dilution, Amersham Life Sciences, Buckinghamshire, UK, PRID: AB_22885189). Image J (RRID: SCR_003070, Rasband, US National Institutes of Health, Bethesda, http://rsbweb.nih.gov/ij, 2010) was used to analyze the images of immunoreactive bands on the membranes.
Statistics
All experimental data are presented in the text and graphs as the mean ± SEM, with an enumerated replicate number (N=x mice/group) in the figure legends. Statistical analysis was carried out using the paired Students t-test, as there were only two groups (VMAT2 Lo mice and age-matched wild-type littermates as the control) to be compared in the present study. It needs to be explained that for the easy writing and space saving, in the following text, legends and figures, we used Ko to represent VMAT2 Lo mice, with an understanding that these mice were 95% knock-down of the VMAT2 gene, rather than real knocking-out. We did not compare the difference between age groups. That means we did not perform the statistics analysis between different ages, for example, comparing the age group of 2 months to the age group of 6 months, or other age groups, although age-related effects for these proteins were observed.
It has been reported that VMAT2 Lo mice showed significantly reduced DA levels in the striatum and cortex from 2 months of age, and in the SN there appeared to be a significantly progressive neurodegeneration with formation of α–synuclein containing inclusions around 18 months of age.31 In addition, starting from 6-months-old these mice also show a progressive degeneration in the LC, and the neuronal number in the LC was significantly reduced at 15 months of age. Furthermore, the degeneration of the SN starts at 18 months of age. Thereby this model replicates important features of PD.23-25 Accordingly in this study, VMAT2 Lo mice at different ages (2, 6, 15, 18 and 23 months) were initially used to examine potential alteration of the DNA repair capacities, and age-matched wild-type littermates were used as the control. However, except for protein levels of OGG1 in the selected brain regions, immunochemical and immunofluorescence labeling showed no significant changes in brain regions of VMAT2 Lo mice at 2, 6, and 15 months of age. Thus, only the measurement results of IHC and IF in mice at 18 and 23 months of age are presented. In addition, four brain regions [the FC, Hip, SN and LC] were selected for measurements. However, although in some brain regions there were some alterations for some measurements between the Ko and control mice, these changes did not reach the statistically significant levels. Therefore, only the data of some brain regions such as the LC and SN are presented.
VMAT2 Lo mice showed an increased protein level of OGG1 in some brain regions
OGG1 is an 8-oxoguanine DNA glycosylase enzyme and expressed mainly in the neuronal cytoplasm.32 It is involved in BER by efficiently recognizing and excising 8-oxo-2'-deoxyguanosine (8-OHdG) presented in DNA damaged by oxidative stress.33 First, the protein levels of OGG1 were measured in the brain regions of these mice by western blotting. As shown in Figure 1A, in the FC of VMAT2 Lo mice at 2, 6, and 15 months of age, OGG1 protein levels were significantly increased, as compared to those of controls (p<0.05, respectively). However, in the FC of VMAT2 lo mice at 18 and 23 months of age, the OGG1 protein levels did not exhibit such changes. On the contrary, the OGG1 protein levels were markedly reduced in the Hip region of VMAT2 Lo mice at 18 and 23 months of age (p<0.05, respectively). Nevertheless, OGG1 protein levels in the Hip region of VMAT2 Lo mice at 2, 6 and 15 months of age did not show significant alterations, either, as compared to those of controls (Figure 1B).
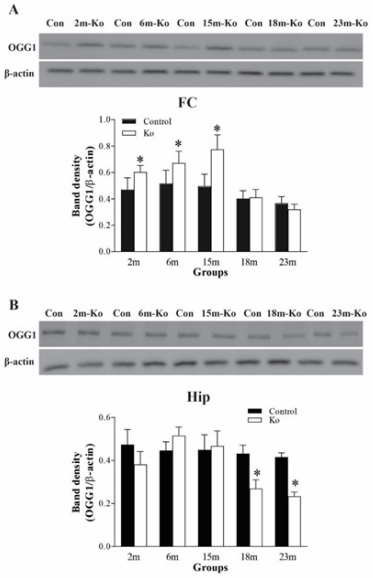
Figure 1 VMAT2 Lo mice have increased protein levels of OGG1 in the FC (A) at 2, 6, and 15 months of age (N=6 mice/group), but have reduced protein levels of OGG1 in the Hip (B) at 18 and 23 months of age (N=6 mice/group). The upper panels in A and B show the representative autoradiographs obtained by western blotting. The lower panels in A and B show quantitative analysis of band densities. * p<0.05, compared to the control. Abbreviations: Con, controls; 2m, 6m, 15m, 18m, 23m, 2-, 6-, 15-, 18-, of 23-month-old mice; Ko, VMAT2 Lo mice.
Figure 2 presents the western blotting-measured results of OGG1 protein levels in the SN and LC. While OGG1 protein levels in the SN from VMAT2 Lo mice at 2 and 6 months of age did not show a significant alteration as compared to those of the controls, a marked elevation of OGG1 protein levels appeared in those mice at 15 months of age (p<0.05). However, similar to those in the Hip, OGG1 protein levels in the SN of VMAT2 Lo mice at 18, and 23 months of age were considerably decreased (p<0.05, respectively, Figure 2A). Unlike those in the SN, there was a significant increase for OGG1 protein levels in the LC in VMAT2 Lo mice at 2, 6, and 15 months of age (p<0.05, respectively). On the contrary, those in the LC in VMAT2 Lo mice at 18 and 23 months of age were markedly reduced (p<0.05, respectively), as compared to those of the controls (Figure 2B).

Figure 2 VMAT2 Lo mice have increased protein levels of OGG1 in the SN (A) and LC (B) at 15 18 and 23 months of age (N=6 mice/group). The upper panels in A and B show autoradiographs obtained by western blotting. The lower panels in A and B show quantitative analysis of band densities. * p<0.05, ** p<0.01, compared to the control. See Fig. 1 for abbreviations.
VMAT2 Lo mice had an increased mRNA level of OGG1 in some brain regions
mRNA levels of OGG1 in the brain regions were measured by qPCR. In contrast to our expectation, mRNA levels of OGG1 in the FC and LC, did not show a significant alteration between VMAT2 Lo mice and controls (data not shown). As shown in Figure 3A, the change of OGG1 mRNA levels in the Hip of VMAT2 Lo mice was similar like that of their protein levels in the same region. That is, while those mRNAs of OGG1 in VMAT2 Lo mice at 2, 6 and 15 months of age did not show marked alteration, those from 18 and 23 months of age exhibited a profound decrease (p<0.05, respectively; Figure 3A). In addition, OGG1 mRNA levels in the SN from VMAT2 Lo mice at 6 and 15 months of age were considerably increased (p<0.05 or p<0.01, respectively), compared to those of controls, and OGG1 mRNA levels in the SN in VMAT2 Lo mice at 23 months of age were meaningfully reduced (p<0.05, Figure 3B).
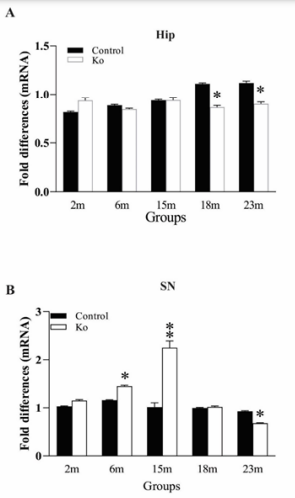
Figure 3 VMAT2 Lo mice have increased mRNA levels of OGG1 in the Hip (A) and SN (B) at 2, 6, 15, 18 and 23 months of age, measured by qPCR (N=5 mice/group). * p<0.05, ** p<0.01, compared to the control. See Fig. 1 for abbreviations.
VMAT2 Lo mice exhibited elevated immunoreactivities of PARP1, ERCC1, XRCC1 and PCNA in brain regions
Besides OGG1, there are also other DNA glycosylases and proteins involved in DNA repair. PARP1 is a nuclear enzyme essential for initiating various forms of DNA repair processes. It is activated in response to genotoxic insults by binding damaged DNA and attaching polymers of ADP-ribose to nuclear proteins at the expense of its substrate NAD+.34 ERCC1 is a structure-specific endonuclease crucial to the repair of DNA damage by the NER pathway.35 XRCC1 is a protein involved in BER and the single-strand break (SSB) repair pathway plays a major role in facilitating the repair of SSBs in mammalian cells, via an ability to interact with multiple enzymatic components of repair reactions.36 PCNA is a DNA polymerase accessory factor that is required for DNA replication during S phase of the cell cycle and for resynthesis during NER of damaged DNA.37 To determine potential alterations of these DNA repair involved proteins in the brain regions of VMAT2 Lo mice, IHC and IF staining were performed. The measurement results indicated that the immunoreactivities of these markers in VMAT2 Lo mice at 2, 6 and 15 months of age did not show significant alterations, as compared to those of controls (data not shown). As shown in the figures presenting the data from mice at 18 and 23 months of age, the density of PARP1 immunoreactivities in the FC (Figure 4A), Hip (Figure 4B), SN (Figure 5A) and LC (Figure 5B) was prominently increased in these brain regions from VMAT2 Lo mice (p<0.01, respectively), as compared to those of the controls. The measurements of ERCC1 in these brain regions are similar to those of PARP1. There were significant increases of ERCC1 immunoreactivities in the FC (Figure 6A), Hip (Figure 6B), SN (Figure 7A) and LC (Figure 7B) (p<0.05 or p< 0.01, respectively), as localized in the nuclei of neurons and astrocytes as reported in the literature.38 A marked elevation of XRCC1 immunoreactivity was only found in the SN (Figure 8A, p<0.01) and LC (Figure 8B, p<0.01), not in the FC and Hip (data not shown).
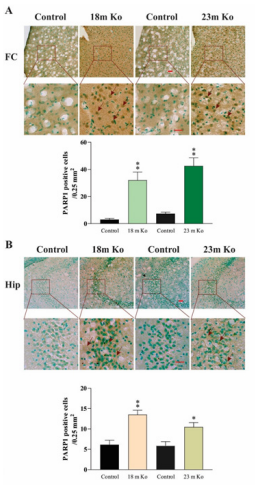
Figure 4 VMAT2 Lo mice showed the immunoreactive levels of PARP1 in the FC (A) and Hip (B), compared with that in age-matched controls, as determined by IHC analysis (n = 6 mice/group). Upper panels in A and B: representative images of IHC staining, and red arrows show positive PARP1 amounts (brown). Lower panels in A and B: subsequent quantification. Scale bar= 25 μm. The values are presented as means ± SEM. * p<0.05, ** p < 0.01 versus the control group. Abbreviations: 18m Ko or 23m Ko, VMAT2 Lo mice at 18 or 23 months of age.
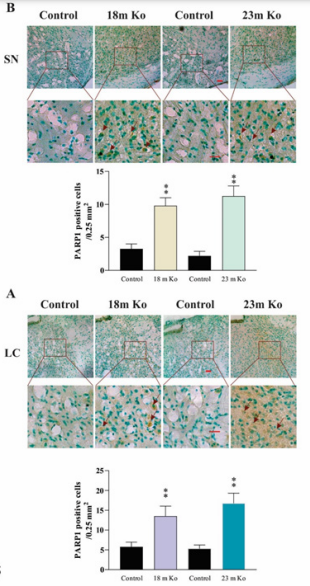
Figure 5 VMAT2 Lo mice showed the immunoreactive levels of PARP1 in the SN (A) and LC (B), compared with that in age-matched controls, as determined by IHC analysis (n = 6 mice/group). Upper panels in A and B: representative images of IHC staining, and red arrows show positive PARP1 amounts (brown). Lower panels in A and B: subsequent quantification. Scale bar= 25 μm. The values are presented as means ± SEM. ** p < 0.01 versus control group. See Fig. 4 for abbreviations.

Figure 6 VMAT2 Lo mice showed the immunoreactive levels of ERCC1 in the FC (A) and Hip (B), compared with that in age-matched controls, as determined by IHC analysis (n = 6 mice/group). Upper panels in A and B: representative images of IHC staining, and red arrows show positive ERCC1 levels (brown). Lower panels in A and B: subsequent quantification. Scale bar= 25 μm. The values are presented as means ± SEM. ** p < 0.01 versus control group. See Fig. 4 for abbreviations.
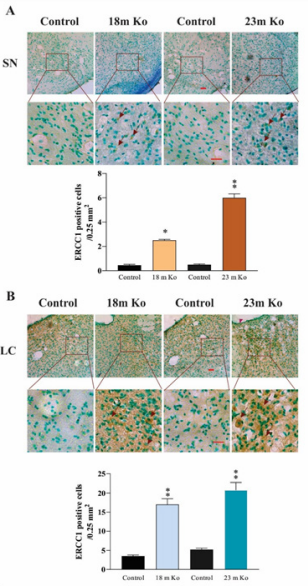
Figure 7 VMAT2 Lo mice showed the immunoreactive levels of ERCC1 in the SN (A) and LC (B), compared with that in age-matched controls, as determined by IHC analysis (n = 6 mice/group). Upper panels in A and B: representative images of IHC staining, and red arrows show positive ERCC1 amounts (brown). Lower panels in A and B: subsequent quantification. Scale bar= 25 μm. The values are presented as means ± SEM. * p<0.05, ** p < 0.01 versus control group. See Fig. 4 for abbreviations.

Figure 8 VMAT2 Lo mice showed the immunoreactive levels of XRCC1 in the SN (A) and LC (B), compared with that in age-matched controls, as determined by IHC analysis (n = 6 mice/group). Upper panels in A and B: representative images of IHC staining, and red arrows indicate positive XRCC1 amounts (brown). Lower panels in A and B: subsequent quantification. Scale bar= 25 μm. The values are presented as means ± SEM. ** p < 0.01 versus control group. See Fig. 4 for abbreviations.
The protein level of PCNA was measured by IF staining. Similarly, compared to those in the age-matched littermate controls, the immunoreactivities of PCNA in the FC, Hip, SN and LC were remarkably increased (Figure 9A) in the VMAT2 Lo mice at the ages of 18- and 23-month-old, which was indicated by quantitative fluorescence image analysis (p<0.05 or p<0.01, respectively, Figure 9B).
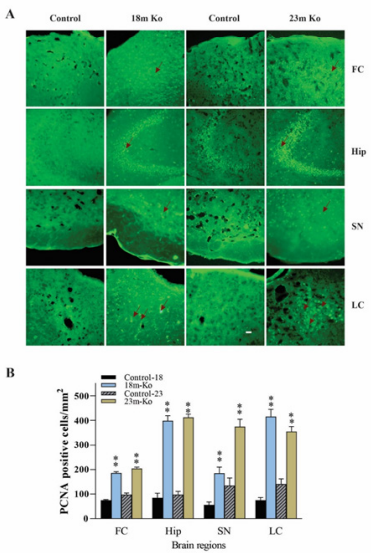
Figure 9 A: VMAT2 Lo mice showed the immunoreactive levels of PCNA by IF staining in the FC, Hip, SN and LC, compared with that in age-matched controls, as determined by IF analysis. Representative photomicrographs of PCNA immunostaining, and red arrows indicate positive PCNA amounts (n = 6 mice/group). B: The graph represents PCNA average positive cells numbers. Representative IF staining to identify PCNA-positive cells with nuclei fluorescently labeled with 4,6-diamino-2-phenyl indole. * p<0.05, ** p < 0.01 versus the control group. Scale bars = 25 μm. See Fig. 4 for abbreviation.
In this paper, we describe experiment results about the DNA repair capacity in the brain regions of VMAT2 Lo mice and controls by measurements of several DNA repair-related enzymes and proteins. The results showed that in the FC and LC of VMAT2 Lo mice at 2, 6, and 15 months of age, OGG1 protein levels were significantly increased. However, the Hip, SN and LC of VMAT2 Lo mice at 18 and 23 months of age exhibited a marked decrease of OGG1 protein levels. These reduced OGG1 protein levels were relatively paralleled by its mRNA levels in these two age groups. Immunochemical staining demonstrated that in the FC, Hip, SN and LC of VMAT2 Lo mice at 18 and 23 months of age there were dramatically elevated immunoreactive densities of PARP1, ERCC1 and PCNA, although such an increase of XRCC1 was only found in the SN and LC from those age groups. These measurement results demonstrated that in these model mice, DNA repair capacities were mostly activated, probably owing to oxidative stress-caused DNA damage. A thorough examination of the DNA damage/DNA repair in this model is of paramount importance in better understanding the critical role of DNA damage and DNA repair in the pathogenesis of PD.
It is well known that OGG1 is a DNA glycosylase with a widespread but heterogeneous distribution among distinct brain regions,39 which repairs the lesion of 8-oxo-7,8-dihydroguanosine (8-oxoG), one of the oxidized forms of guanine in DNA.40 Once 8-oxoG has formed in DNA, OGG1 removes this oxidized base and produces a 3’ nick to initiate BER. At least two isoforms of human OGG1 proteins encoded by alternatively spliced mRNAs differentially contribute to the BER pathways for 8-oxoG generated in both nuclear and mitochondrial DNAs.33 It was reported that 8-oxoG lesions were accumulated in OGG1 knockout mice.41 In addition, OGG1 also possesses apurinic/apyrimidinic (AP) lyase activity.32 Recent reports demonstrate that OGG1 is neuroprotective in neurotoxicological paradigms that impact the nigrostriatal.42,43 Even if there is a direct oxidation of guanine in DNA, OGG1 can efficiently repair the oxidized base.33 In the present study, altered protein and mRNA levels of OGG1 generally exhibited a biphasic pattern. For example, the FC and LC of VMAT2 Lo mice at the age of 2-, 6- and 15-months-old showed an increased OGG1 protein level. However, in the Hip, SN and LC of VMAT2 Lo mice at 18 and 23 months of age, OGG1 protein levels were significantly diminished, which was accompanied by a reduction of OGG1 mRNAs in the Hip and SN from the same age group. An increased OGG1 level in these brain regions is somewhat consistent with the previous report of western blot analysis that the levels of OGG1 in the SN of PD brains was 1.6-2.9-fold higher than that of the age-matched control.13,44 As for the reduced level of OGG1 in aged model mice one possible explanation is the age factor, because OGG1 levels in various brain regions in mouse45,46 and human brains47 were reported to be steadily decreased with increasing ages. Another possible explanation is that in the younger model mice in present study the severe DNA damage reaction triggers an upregulation of OGG1, then in older model mice the capacity of the OGG1 to repair oxidatively damaged DNA has been diminished, exhibiting as a reduction level. In addition, OGG1 activity and amount are inversely related to the extent of oxidative DNA damage.48 Coincident with this, it was reported that those who had PD for longer than 10 years did not exhibit intense cytoplasmic immunostaining of OGG1, compared to an increase of OGG1 level in PD patients with a duration of less than 10 years.13
For such biphasic patterns of OGG1 levels at different ages, there may be one possible explanation by in vitro experiments.49,50 For example, it was reported that DNA repair, represented by enzymatic activity of OGG1, was upregulated by about 20% in response to a 7-fold increase in 8-oxodG in cells that were incubated with single toxicants and their combinations for 24 h. However, at the third day of incubation, the OGG1 activity dropped significantly below control levels.51 Noticeably, at that time after incubation with a single or combination of toxicants, superoxide dismutase activity remained elevated over control levels. Interestingly, western blot measurements showed that after 24 h of incubation these treatments did not alter the actual amount of OGG1 protein. It suggested that the treatment with the toxicants only altered enzymatic activity of OGG1 rather than the protein levels. However, after 72 h of incubation, OGG1 protein levels did decrease. Such diminished OGG1 level may be explained as an increase in apoptosis in these PC12 cells in their differentiated state compared with their proneuronal proliferative state.52 In addition, our data about OGG1 level alterations in VMAT2 Lo mice do not seem to be consistent at different brain regions. It may be due mainly to brain regions-specific variation in DNA BER activity.53 Also, the DNA repair response to an oxidative challenge varies across brain areas. For example, the midbrain has the highest levels of oxidative DNA lesion after being exposed to an oxidative challenge, and its upregulation of OGG1 capacity is least in this respect.48
In the present study, the immunoreactivity of PARP1, ERCC1, XRCC1 and PCNA in the brain of VMAT2 Lo mice was examined. To select these enzymes and proteins is based on following facts. PARP1 is one of 18 members in the PARP family and catalyzed post-translational protein modification to regulate other cellular proteins related to DNA repair. PARP1 is not only involved in both BER and DSBR,54 but also appears to be involved in NER.55 Generally, DNA damage triggers activation of the PARP1 to recruit DNA repair proteins and interacts with core NER proteins to fix the damaged DNA. ERCC1 is a structure-specific endonuclease, and a rate-limiting component in the NER pathway.35 It also involved in DSBR and interstrand cross-link repair.56,57 In addition, ERCC1 plays a major role in facilitating the repair of SSBs in mammalian cells.58 For example, both the global and neuron-specific Ercc1 mutant mice developed signs of genotoxic stress and mild neuronal degeneration during adult life, indicating that unrepaired DNA damage is sufficient to cause progressive neuronal pathology and neuronal plasticity deficits.59 Accordingly, it is an essential DNA repair enzyme in the brain. XRCC1 is a protein involved in BER and the SSB repair pathway, and plays a major role in facilitating the repair of SSBs in mammalian cells, via an ability to interact with multiple enzymatic components of repair reactions.36 There are high levels of 8-OHdG in XRCC1 mutant genotypes.17 PCNA is a DNA polymerase accessory factor that is required for DNA replication during S phase of the cell cycle and for resynthesis during NER of damaged DNA.37
In the present study, a striking characteristic of all of these four protein measurements is their increased immunoreactivity in measured brain regions of VMAT2 Lo mice at 18 and 23 month of ages. While the observations of upregulated levels of PARP1 and XRCC1 is coincident with the previous report of a significant elevated level of PARP1 and XRCC1 in the SN of PD patients,17,60 the similarly upregulated levels of ERCC1 and PCNA can be found from animal studies. For example, ischemia can lead to DNA damage, and brain ischemia usually induces specific expression of genes related to the DNA repair pathways. It was report that in a rat PD model with 6-hydroxydopamine, treatment with an agonist 7-OH-DPAT resulted in significant increases in PCNA labeling and their induction in the SN pars compacta at all-time points examined.61 As PCNA is required for DNA synthesis and NER, the elevated PCNA as a marker of cell cycle and endogenous cell proliferation reflects the agonist-induced repair in injured cells.62 In addition, PCNA level was upregulated in the Hip following global ischemia.63 Moreover, ischemia markedly induced long-lasting BER activity by upregulating protein levels and activity of essential BER enzymes including XRCC1,64 and ischemia also increased ERCC1 at 3 days, remaining until 14 days in the cortex and striatum.38 Taken together, obviously these increases may have some indications. First, increased activation of these enzymes and proteins reveals the increased DNA damage in VMAT2 Lo mice. For example, 8-OHdG lesion was found to be high in PD patients compared with controls having XRCC1 mutant genotypes. Induced BER activity constitutes an important endogenous mechanism that protects the brain against insults such as ischemia-induced oxidative neuronal injury. Second, increased levels of DNA repair factors can directly contribute to enhanced endogenous DNA repair activity and neuroprotective effects. It was reported that ERCC1 is capable of protecting neurons against ischemic injury in the rat brain and ERCC1 over-expression significantly reduced DNA damage and infarct volume.64,65 Generally, cellular DNA damage triggers activation of the PARP1 to recruit DNA repair proteins and fix the damaged DNA. In addition, activated PARP1 interacts with other core NER proteins and may play a role in maintaining mtDNA integrity.66 Supplementation with PARP1 inhibitors extends life span in wild-type and DNA repair–deficient groups.67,68 PARP1 deletion or inhibition showed protective effects in PD animal models. In addition, the activation of PARP1 directly caused the aggregation of α-synuclein in PD neurons.69,70
The present study used different methods to examine the alteration of different DNA repair-related enzymes and proteins in VMAT2 Lo PD model mice. The results demonstrated that OGG1 level seems to have a biphasic alteration between relatively younger and aged mice. Furthermore, there was an upregulation of PARP1, ERCC1, XRCC1 and PCNA in the different brain regions. The consequence of these upregulated DNA repair-related proteins indicates that in this mouse PD model, some DNA repair processes were activated to maintain genomic integrity. Accordingly, these data underscore the importance of DNA repair capacity in the initiation and progress of development of PD and indicates the feasibility of VMAT2 Lo mice in the investigation of PD.
We have read and have abided by the statement of ethical standards for manuscripts submitted to Neuroscience. Informed consent was obtained from all individual participants included in this study. All experiments were authorized by the Institutional Animal Ethics Committee of East Tennessee State University, and procedures were performed in compliance with the Guide for the Care and Use of Laboratory Animals.
The authors declare that there is no conflict of financial or non-financial interest that are directly or indirectly related to the work submitted for publication.
This work is supported by NIH grant AG055107.
The datasets used and/or analyzed during the current study available from the corresponding author on reasonable request.
All authors had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis. M.Y. Zhu: designed and conceived study, acquired and interpreted the data, drafted the manuscript. K. Parker; F. Zeng F; YQ Zhan, M. Miller: investigation and analysis. K. Parker and M.Y. Zhu: revision and editing.

©2023 Parker, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.