MOJ
eISSN: 2374-6920


Short Communication Volume 3 Issue 6
Biotech Support Group, USA
Correspondence: Swapan Roy, Biotech Support Group, 1 Deer Park Drive, Suite M Monmouth Junction NJ 08852, USA, Tel 7322742866
Received: July 27, 2016 | Published: August 5, 2016
Citation: Roy S, Kuruc M. The functional subproteomes of serpin protease inhibitors are now open for LC-MS biomarker discovery. MOJ Proteomics Bioinform. 2016;3(6):152-155. DOI: 10.15406/mojpb.2016.03.00106
Conformational variants of the unique family of protease inhibitors annotated as SERPINs, are most often underrepresented in proteomic analyses. This limits understanding the complex regulation that this family of proteins presents to the networks within the protease web of interactions. Using bead-based separation provided by the NuGel™ family of proteomic enrichment products - notably AlbuVoid™ & AlbuSorb™, we demonstrate their utility to satisfy investigations of serum SERPINs. We also suggest their use to develop functional profiles of the SERPIN Proteoform, and how those can establish relationships to disease phenotypes, gene mutations, and deregulated mechanisms.
The balance and regulation of proteolytic activity within serum is essential to blood based biomarker discovery and possibly to therapeutic intervention. Changes in blood components often reflect acute responses to thwart external stresses, such as coagulation when skin is severed, or inflammatory response during microbial infections. These fast-acting responses are controlled by proteolytic cascades, essentially modifying functionality by the controlled degradation of protein structures. While necessary for acute response, persistent activation of these proteolytic cascades can lead to chronic conditions. So, there is a balance and regulation of these proteolytic cascades which is necessary to keep aberrant proteolysis controlled (Figure 1).
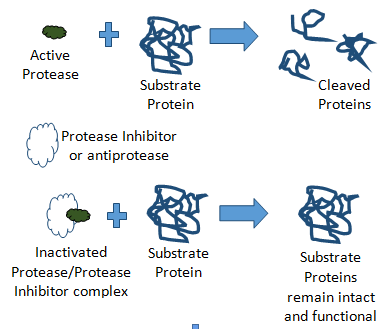
This is done through systemic regulatory protein factors, called protease inhibitors or antiproteases. What is now becoming apparent is that the influence of inhibition can be just as important as zymogen activation in rapid switch cascades controlling subnetworks within the protease web.1 One such example is the Alpha-1-Antitrypsin (the inhibitor) substrate activation of proMMP-2, a metalloproteinase involved in tumor invasion and angiogenesis.2
It is therefore necessary to consider that inhibitors are themselves being regulated under different and often complex means of regulation. Within this context, therein lies the special case of the SERPIN superfamily of protease inhibitors.
There are many different families of protease inhibitors, one exceptional family being the serine protease inhibitors, a collection of a super-family of proteins annotated within the SERPIN gene nomenclature. These proteinase inhibitors regulate key intracellular and extracellular pathways. Among the key regulators in blood serum, SERPINA1 (also known as ɑ1-antitrypsin) protects lung tissue from neutrophil elastase, SERPINC1 (also known as antithrombin) controls coagulation proteases, SERPING1 (also known as plasma C1 inhibitor) regulates complement activation, and SERPINF2 (also known as ɑ-2-antiplasmin) inhibits plasmin and regulates fibrinolysis.2,3
The role of the SERPINs in these critical functions is rarely straightforward as is the case for more simplistic binary binding inhibition. Unlike binary binding and associations, SERPIN functional regulation is not linear with respect to differentiating phenotypes based on strict abundance measurements, such as data derived from ELISA or quantitative LC-MS. An aggregate of all sub-populations does not account for important differences amongst the many sub-populations generated by conformational variants within this superfamily of proteins (Figure 2). This is because SERPINs differ from all other families of protease inhibitors in having a complex mechanism of action that involves a drastic change in their shape, forming the basis of a suicidal substrate inhibition mechanism.2,3 The reactive centre loop (RCL) extends out from the body of the protein and directs binding to the target protease. The protease cleaves the serpin at the reactive bond site within the RCL, establishing a covalent linkage between the carboxyl group of the SERPIN reactive site and the serine hydroxyl of the protease.4 The resulting inactive serpin-protease complex is highly stable, and the structural disorder induces its proteolytic inactivation. As a consequence, the protease is permanently inhibited and functionally inactivated. Nevertheless, the story does not end there for the inhibitor, as after the initial interaction with the substrate protease, one of two possible outcomes can occur.
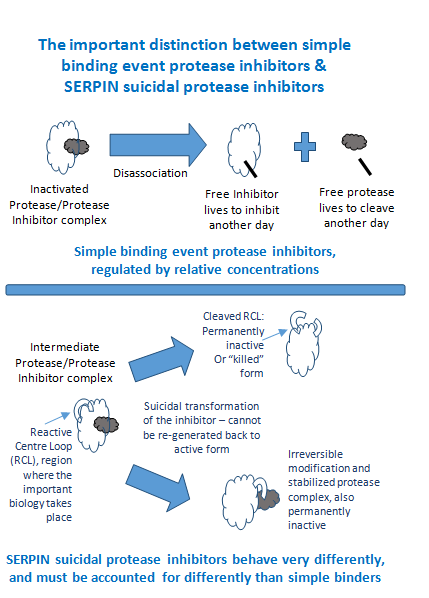
One possible outcome is driven by covalent modification permanently inactivating the inhibitory capacity as the SERPIN peptide reactive bond region is irreversibly bound to the protease, and thus cannot be reconstituted back to an active form. The second possible outcome is a permanently inactive or “killed” Proteoform of the SERPIN as the peptide RCL region is cleaved and can no longer bind target substrates.4
As a result, even minor changes in the structure due to genetic variation and post-translational modifications, can modify the function of SERPINs and give rise to a variety of clinical presentations. Some 200 different mutations in serpins are known to result in disease.5 In particular, mutations affecting antithrombin confer a predisposition to thrombosis, those affecting C1 inhibitor confer a predisposition to angioedema, and those affecting antiplasmin confer a predisposition to hemorrhage. Interestingly, an alternative function is made possible by a mutation in which the methionine in the RCL region of Alpha-1-Antitrypsin is replaced by an Arginine converting its function as an inhibitor of neutrophil elastase to a highly effective inhibitor of the coagulation proteases; the consequence of which is life-threatening hemorrhagic disease.6
Mutations can affect function throughout the sequence. However, the most common loss of serpin function from mutation, are those affecting the mobile hinges of the molecule within or near the RCL. These lead to spontaneous changes in conformation that allow either the insertion of the intact reactive loop into the main β sheet, resulting in the formation of an inactive “latent” form, or the insertion of the loop of one molecule into the β sheet of the next, resulting in the formation of polymers. Polymerization occurs in alpha 1-antitrypsin with the common Z variant mutation, leading to decreased secretion from the liver into the circulation, resulting in emphysema and cirrhosis.7 Amino-acid substitution in the RCL region is the likely event transforming the non-inhibitory serpins. Post-translational modifications at the RCL region such as oxidation of methionine in Alpha-1-Antitrypsin have also been proposed as a source of dysfunction.8
So understanding the underlying mechanisms, contributions from genetic wiring or environmental stresses, and their relationships with aberrant proteolysis are necessary to characterize disease. Proteomic analyses offer a new lens of observation to examine the resulting conformational variants that can be reported as potential biomarkers of disease phenotypes. As an example, one such inhibitor SERPINA1, known more commonly as Alpha-1-Antitrypsin (AAT), has several isoforms observed in plasma using 2-DE, and often serves as a model for conformational diseases.5,9 Circulating levels of AAT are between 1.2 and 2mg/ml in healthy persons, but are known to increase during acute phases of inflammation and infection. Its function and activity is controlled by the many variants attributable to its conformational nexus of features; the term ‘proteoform’ is often used to describe such conformation features and we adopt that term here.
Several reports observe that the conformational properties of AAT have multiple effects on tumor cell viability and diverse roles in tumorigenesis, suggesting such isoforms may display a specific basis for diagnosis of cancer and neurodegenerative disorders.8,10,11 Yet, most often in proteomics, all sub-populations of AAT are rolled into and counted as one homogeneous population, or as in the case of immuno-depletion, simply ignored as background noise. As a result, the regulation, balance and dynamism within these systems and its impact on the protease web of disease progression cannot be properly investigated.
By combining our unique strategies of binding and voiding high abundance proteins, we can observe different sub-populations with characteristic binding biases. We have previously reported for Alpha-1-Antitrypsin, that the resultant cleaved-RCL proteoform, and the uncleaved-RCL proteoform are very distinctive sub-populations, separated by AlbuVoid™, and reported at the peptide feature level by LC-MS.12 In this report, we consider how our Albumin Removal products - AlbuVoid™ and AlbuSorb™ (Biotech Support Group LLC, Monmouth Junction NJ USA), can help to functionally profile and unravel this complex world of the SERPIN superfamily of proteins.
Through a proprietary polymer coating, 50µm porous silica beads are crosslinked and passivated. This is the foundation of the NuGel™ surface chemistry. From this foundation, a library of bead architectures has been created. Each bead chemistry in the library presents a singular mixed-mode interaction; combining elements of ionic, aliphatic and aromatic hydrophobicity, and polymeric characteristics. One can think of these binding interactions in different terms; as general non-specific protein adsorbents, or as bead matrices with weak affinity or imperfect fit interactions. In this way, their binding behavior is very different from classical high affinity binding which demands near perfect fits. Under protein saturation conditions, progressive displacement provides a separation bias towards or against select proteins. As a result, all derivative NuGel™ products were empirically characterized to meet the needs of the application; for example, AlbuVoid™ to selectively void (not bind) Albumin with special bias towards the vast majority of the remaining low abundance serum proteome on the bead. Two NuGel™ based products support Albumin Removal:
In this model case, the ratio of the ACTIVE sub-population vs. the INACTIVE sub-population is greatly altered in disease, whereas simple abundance measurements of the total population would not be very informative. In the following Figure 4, we report on the SERPINs observable by LC-MS and how they bias towards AlbuVoid™ and AlbuSorb™ (Table 1).
Protein |
Also Known As |
Function |
AlbuVoid™ Bead Bound S.Cts. |
AlbuSorb™ Flow through |
Reactive bond site14 |
Notable variants14 |
SERPINA1 |
Alpha-1-Antitrypsin (AAT) |
Inflammation, elastase inhibition |
59 |
519 |
Met382-Ser383 |
Z Variant {Glu366→Lys366} deficiency syndrome, Pittsburgh variant {Met382→Arg382} life-threatening bleeding |
SERPING1 |
Plasma Protease C1 inhibitor |
regulates complement cascade, levels rise ~2-fold during inflammation |
51 |
63 |
Ala465-Arg466 chymotrypsin, Arg466-Thr467 |
|
SERPINA3 |
Antichymotrypsin |
Apoptosis, Alzheimer’s, inflammation |
86 |
117 |
Leu383-Ser384 |
|
SERPIND1 |
Heparin cofactor II |
Coagulation, Thrombin inhibitor activated by Heparin |
124 |
28 |
Leu463-Ser464 |
|
SERPINA8 |
Angiotensinogen (AGT) |
angiotensin I precurser, blood pressure regulation, non-inhibitory |
4 |
62 |
none |
disulfide bond is labile, near 40:60 ratio with the oxidized disulfide-bonded form |
SERPINC1 |
Antithrombin, ATIII |
inhibits thrombin, regulates coagulation, angiogenesis, heparin cofactor |
58 |
79 |
Arg425-Ser426 |
mutations/variants can lead to increased risk of thrombosis, alter functional heparin & thrombin binding domains |
SERPINF1 |
Pigment epithelium-derived factor, PEDF |
Neurotrophic factor, non-inhibitory |
45 |
0 |
none |
|
SERPINA4 |
Kallistatin |
Kidney function, inflammation |
45 |
0 |
Phe388-Ser389 |
cleavage at the reactive site by tissue kallikreins |
SERPINF2 |
ɑ-2-antiplasmin |
Fibrinolysis, inhibitor of plasmin and trypsin |
10 |
39 |
Arg403-Met404 plasmin, Met404-Ser405 chymotrypsin |
Alanine insertion at the reactive site promotes serious bleeding disorders |
SERPINA10 |
Z-dependent proteinase inhibitor |
Coagulation regulation |
23 |
0 |
Tyr408-Ser409 |
Tyr408→Ala408 loss of inhibition |
SERPINA5 |
Protein C inhibitor |
Coagulation, inflammation |
13 |
0 |
Arg373-Ser374 |
Variants near or at the reactive bond alter inhibition of thrombin activity |
SERPINA6 |
Corticosteroid-binding globulin |
Hormone transport, non-inhibitory |
0 |
26 |
none |
|
SERPINA7 |
Thyroxine-binding globulin |
Hormone transport, non-inhibitory |
0 |
17 |
none |
|
Table 1 SERPINs observable by LC-MS
We believe that conformational changes associated with the cleavage of the reactive bond, confer more or less binding affinity to the non-specific interactions with our beads. Such cleavage stabilizes protein structures; AlbuVoid™ binding especially biases towards unstructured proteins, for example AAT RCL-intact proteoform binds favorably over the RCL-cleaved proteoform.12 Noteworthy is that several non-inhibitory SERPINs A6-8, all bind poorly to AlbuVoid™, supporting evidence for the role of conformational stability in binding biases.
By using bead-based enrichment tools, investigators can now develop relationships between the proteoforms presented by genetic variation, environmental stresses, post-translational modifications, and disease phenotypes. By using the peptide reporting features of the RCL peptide regions within SERPIN inhibitors, both “active” and “inactive” proteoforms are now distinguishable, adding a new level of proteomic characterization to the underlying mechanisms of disease. Bead-based proteomic enrichment products as described can support the functional and structural proteomic analyses necessary to characterize these conformational sub-populations so that they may become useful biomarkers for disease.
None.
The author declares no conflict of interest.

©2016 Roy, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.