MOJ
eISSN: 2374-6920


Research Article Volume 3 Issue 5
1Department of Biotechnology, Bapuji Institute of Engineering and Technology, India
2Savi Easy Life, Czech Republic
Correspondence: Sreenivas Reddy Bathula, Department of Biotechnology, Bapuji Institute of Engineering and Technology, Davanagere - 577004, Karnataka, India,, Tel +919591240234
Received: May 01, 2016 | Published: June 20, 2016
Citation: Bathula SR. Structural comparison between Trp-cage and retro Trp-cage peptide structures. MOJ Proteomics Bioinform. 2016;3(5):116-122. DOI: 10.15406/mojpb.2016.03.00097
Ab-initio protein folding prediction still remains a challenging task despite an increase in computational power. To understand details of the protein folding we reverted the Trp-cage miniprotein and used it for in silico predictions. We also solved its nmR structure. Amino acid contacts in the predicted models do not match the experimental Trp-cage or retro Trp-cage. The investigation revealed few similarities in crucial contacts of nmR experimental structures of both peptides. None of the tested servers could predict correct conformation of disorder-promoting residues.
Keywords: Trp-cage, retro Trp-cage peptide, nmR, ab-initio, poly-proline type ii, trifluoroethanol, phosphate buffer, tyrosine, tryptophan
PPII, poly-proline type ii; TFE, trifluoroethanol; PB, phosphate buffer; TC5b, trp-cage forming
The 20 residue Trp-cage miniprotein is an ideal model for experimental studies of protein folding mechanisms and computer simulations involving molecular dynamic studies.1,2 The miniprotein consists of an order-promoting3 α-helical part made up of residues mostly involved in fold stabilization and a disorder-promoting4 poly-proline type II (PPII) helical part with residues involved in Trp burial.5 The significance of individual amino acid residues inside the Trp-cage was studied by mutations at various locations. We were interested in the nature of the folding of structural motifs and their inter-residual interactions. Therefore, instead of adopting a strategy using individual mutations in alpha-helices and in PPII-helices, we completely reversed the peptide sequence from N-terminal to C-terminal and studied the folding in detail and also prepared mutants (P9A and P9S) on the retro peptide (Table 1). These two mutations occur in the 310-helix / beta-turn forming region, where residue folding is critical for making contact between N and C terminals. The retro peptide backbone is similar to the regular miniprotein in the alpha-helices and PPII-helices part. The mutations in the retro peptide do not change this behaviour. However none of the retro peptide is able to form a proper Trp-cage
A peptide with reversed sequence (with respect to the original Trp-cage) was investigated to test the hypothesis that the Trp-cage fold is resistant to the retro operation. CD spectroscopy showed that the peptide was unstructured in water but formed a α-helical structure in 30 % Trifluoroethanol (TFE) at 2 ˚C. Its 3D structure was determined using distance restraints derived from 2D NOESY spectra.
Truncation and mutation of the folded part of the 39-residue exendin-4 peptide produces a 20-residue folded Trp-cage. We reversed and mutated the folded peptide and studied their folding.
Retro Trp-cage peptides were synthesised chemically by C-terminal amidation. Peptide structure analysis was done by CD and NMR spectroscopy. The methods are discussed in detail.24
CD spectra and melting studies
Well constructed retro Trp-cage peptide and its mutants were dissolved in 15 mM phosphate buffer (PB), pH 7. To measure CD spectra, 66 micromolar (μM) samples were prepared both in the presence and the absence of trifluoroethanol (TFE). Melting temperature calculation, helical analysis, temperature refolding, and TFE effect were studied by CD spectra. CD spectra were recorded at 2 °C using a Jasco J-810 spectrometer (Jasco, Japan) equipped with peltier cell holder. Data was collected from 185 to 260 nm, at 100 nm/min, 1s response time and 2 nm bandwidth using a 0.1 cm quartz cuvette containing the protein sample (Table 2). Each spectrum was the average of ten scans and was corrected for absorbance of the buffer. Collected CD data was expressed in terms of the mean residue ellipticity (ΘMRE) using the equation:
where Θobs is the observed ellipticity in degrees, Mw is the protein molecular weight, n is the number of residues, l is the cell path length, c is the protein concentration and the factor 100 originates from the conversion of the molecular weight to mg/dmol.
Exendin-4(PDB ID: 1JRJ) HGEGTFTSDLSKQMEEEAV RLFIEWLKNG GPSSGAPPPS-NH2 |
TC5b (Trp-cage PDB ID: 1L2Y) NLYIQWLKDG GPSSGRPPPS |
Retro Peptide (PDB ID: 2LUF) SPPPRGSSPGG DKLWQIYLN-NH2 |
Retro Peptides Mutants (P9S & P9A): SPPPRGSSSGG DKLWQIYLN-NH2 |
SPPPRGSSAGG DKLWQIYLN-NH2 |
Table 1 Peptide sequences related to Trp-cage
Peptide |
n |
Mw (Da) |
c (mg/ml) |
Solvent |
P9A |
20 |
2143.38 |
0.1333 |
PB |
P9A |
20 |
2143.38 |
0.1333 |
PB + 30 % TFEa |
P9S |
20 |
2159.38 |
0.1499 |
PB |
P9S |
20 |
2159.38 |
0.1499 |
PB + 30 % TFEa |
Retro Peptide |
20 |
2169.41 |
0.1431 |
PB |
Retro Peptide |
20 |
21.69.41 |
0.1431 |
PB + 30 % TFEa |
Table 2 Studied variants of retro Trp-cage miniprotein and its mutants
aTFE – trifluoroethanol
NMR sample preparation and structure ensemble generation
All samples including mutants were prepared for NMR measurements in a total volume of 600μl. The solution contained 1mM peptide and 30% TFE, 10% D2O, 0.03% NaN3, rest 60% 15 mM phosphate buffer at pH 7. All samples were prepared from ~1.3 mg of lyophilised protein. At higher concentrations (>1mM), quick aggregation/association was observed. Samples of 1mM concentration survived for 3 days at low temperatures without any aggregation/association. Samples were temperature refolded every time before measurement.6,7 The samples slowly solidified, indicating that solvent interactions with peptide decreased over time.8
NOESY, TOCSY, COSY, and 1H-13C HSQC spectra were measured using a 500 MHz Bruker Avance III NMR spectrometer equipped with a TXI probe. P9A NOESY spectra were measured using a 600 MHz Bruker Avance III NMR spectrometer equipped with the TCI CryoProbe. All the spectra were processed using the NMR Pipe software. The TOCSY (mixing time 50ms) and COSY spectra were used to determine intra residual correlations. NOESY spectra recorded with a 200 ms mixing period was used to determine the sequential connectivity and to get the final structure. Software programs Sparky (T.D. Goddard and J. Kneller, University of California, San Francisco) was used for peak picking and assignments, Aria version 2.1 for NOE assignment and CNS version 1.2 for structure calculations, Record scripts for structure refinement. After CING validation, NMR structures were deposited in the RCSB data bank.
CD spectra analysis
Helix formation is a prerequisite for proper Trp-cage formation. Normally, for such short peptides, helical folding is quite difficult (Figure 1). I used 30 % TFE to induce helix formation. CD spectra (Figure 2) show helix formation following addition of TFE. To further study the TFE effect, the percentage of TFE increased to 50 %, but there was no further change in the helical content. Trp-cage makes helices in normal phosphate buffer without TFE, but for the reverse sequence, we need to induce it using a chemical agent. Trp-cage melting temperature is 43.9 ± 0.8 °C in aqueous buffer at pH 7.10,11 The CD spectra showed that retro Trp-cage in 30% TFE has a melting temperature of 31.7 ± 0.6 °C (Figure 3). Their mutants showed almost similar stability. This tells us that high melting temperature is due to hydrophobic bond contributions to cage formation.9
The retro peptide mutants P9A and P9S showed CD spectra similar to the unmutated retro peptide both in the presence and absence of TFE (Figure 4). The mutations increased the flexibility of the 310-helix / beta-turn forming region. Structural studies were done on these mutants, but there was no cage formation observed in these peptides (Figure 10). Peptide samples were heat refolded before NMR measurements. Therefore, it was interesting to observe how the peptide behaves at higher temperatures. Figure 5 shows peptide CD spectra measured at 2 °C in the presence of 30 % TFE. The samples were then rapidly heated to 84.5 °C and rapidly cooled to 2 °C. There was no change observed before and after refolding in CD spectra. Helical content increase was observed while heating in the presence of 30 % TFE.

Figure 1 Far-UV CD spectra of retro Trp-cage miniprotein at various temperatures in phosphate buffer at pH 7, spectra from 2 °C (black) to 75 °C (red) with selectable set points in incrementsof 1 °C/min only shows disordered like signature.
NMR analysis
Retro peptides were pre-checked by CD spectroscopy before the NMR analysis to determine whether the peptides were folded and stable. Initially, NMR samples were prepared only with phosphate buffer. 1D spectra were measured and TFE was added to sample and 1D spectra were measured again (Figure 6). The change observed in the spectra showed a TFE effect on retro peptide folding.

Figure 2 Far-UV CD spectra of retro Trp-cage mini protein in the presence of 0%, 30% and 50% TFE. All measurements were done at 2 °C.

Figure 3 Far-UV CD spectra of retro Trp-cage mini protein in the presence of 30% TFE at different temperatures from 2 °C (black) to 75 °C (red) with selectable set points in increments of 1 °C/min. Determined melting temperature (Tm = 31.7 ± 0.6 °C) of retro Trp-cage mini protein in the presence of 30 % TFE.
NMR-structure determination of the retro Trp-cage peptides
Inside the peptide each distinct nucleus has a distinct chemical environment. Chemical shifts can be evaluated for individual nuclei using multidimensional experiments. The 2D TOCSY, COSY and NOESY experiments were used in this study to assign the observed frequencies of the corresponding nuclei of retro Trp-cage and its mutants and to provide data for structure calculation. The NMR experiments usually started by excitation of pulses of electromagnetic waves, the mentioned pulse sequences allowed us to investigate and select specific types of correlations between nuclei. In TOCSY and COSY experiment, magnetization was transferred through the chemical bonds, while the transfer through space, irrespective of the covalent structure was utilized in NOESY. The TOCSY was used in combination with NOESY to assign the different chemical shifts to specific nuclei (Figure 7), and NOESY was used to generate the distance restraints used in the structure calculation (Figure 8) of unlabelled retro Trp-cage peptide and its mutants.
The spin systems of the individual amino acids were identified in the 2D TOCSY and COSY spectra. The TOCSY shows off diagonal cross peaks between all protons in the spin system, but COSY exhibits only cross peaks between neighbours. Proton resonance frequencies were assigned using the traditional approach based on 2D NOESY and TOCSY spectra. Only one set of peaks was observed in the spectra, indicating that a majority of the mini-protein is present in one folded form at 30 % TFE concentration.
The NOESY experiment transfers magnetization through space, and shows cross peaks for all protons that are close in space (closer than 5 Ǻ) regardless of whether they are in the same spin system or not. Sequential assignment was based on the observed short-range Hα i-HN i+1 NOESY cross-peaks and confirmed by sequential NOEs between side-chain protons. Medium-range and HN-HN i+1 NOEs were used to determine which residues form the alpha helical structure. The neighbouring residues are inherently close in space, so the assignments can be made based on the peaks in NOESY with other spin systems (Table 3). The algorithm converts the restraints and the general protein properties into energy terms. The process results in an ensemble of structures corresponding to the fold of the retro Trp-cage peptides. A set of 300 structures was calculated in vacuum and among them, 100 structures with the lowest energy were further refined in an explicit water solvent.
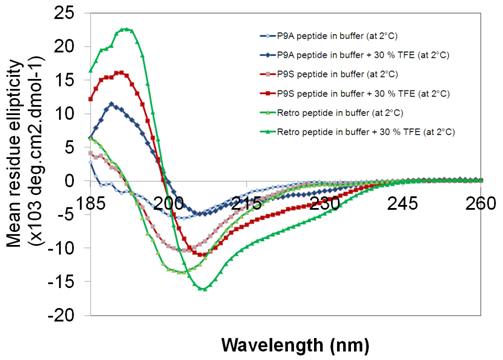
Figure 4 CD spectra data of retro Trp-cage peptide and mutants (P9A and P9S) both in the presence and absence of TFE.

Figure 5 Far-UV CD spectra of retro Trp-cage mini protein in the presence of 30 % TFE measured at 2°C before thermal unfolding and refolded after thermal denaturation. Retro peptide CD spectra measured at 2 °C (shown in red) in the presence of 30 % TFE then quickly heated up to 84.5 °C (shown in blue) and again cool it down to 2 °C (shown in green).

Figure 6 Proton chemical shift positions of retro peptide in phosphate buffer (shown in red) after addition of 30 % TFE (shown in blue) measured at 2 °C.

Figure 7 Retro peptide fingerprint region of the TOCSY spectrum recorded with a 200 ms mixing time, shows inter residual correlations.
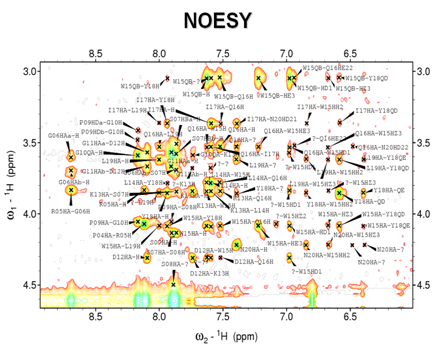
Figure 8 Retro peptide fingerprint region of NOESY spectra showing sequential connectivity due to dipolar interactions among nuclei in 5 Ǻ spatial proximity.
Assignment |
w1 |
w2 |
Data Height |
Note |
R05QG-W15HD1 |
1.134 |
6.977 |
872291 |
1 |
R05HBa-W15HD1 |
1.238 |
6.978 |
833650 |
1 |
P04QG-P09HDb |
1.661 |
3.482 |
5757164 |
1 |
P09HDb-P04QG |
3.482 |
1.660 |
5667692 |
1 |
S01HBb-L14HBb |
3.549 |
1.443 |
1217150 |
1 |
S01HBb-L14HG |
3.550 |
1.327 |
967917 |
1 |
G11HAa-P03QG |
3.564 |
1.653 |
1889681 |
1 |
K13HA-S07H |
3.844 |
7.865 |
1491629 |
1 |
N20HA-W15HZ3 |
4.216 |
6.673 |
1475014 |
1 |
S08HA-I17HB |
4.492 |
1.634 |
1389023 |
1 |
W15HH2-N20HBb |
6.846 |
2.455 |
1515053 |
1 |
W15HH2-N20HBa |
6.849 |
2.419 |
1235989 |
1 |
Table 3 Starting set of long-range NOEs of the retro Trp-cage
Structure validation
The quality of a subset of 10 structures with the lowest energy was validated using the CING program and deposited in the Protein Data Bank database under the accession code 2LUF (Figure 9). The structure determination statistics are summarized in.24
The C-terminal part of all studied peptides forms a helix (Gly11-Gln16) approximately corresponding to the N-terminal Trp-cage helix. Its overall similarity to the Trp-cage structure could be captured by a Cα superposition with a Cα RMSD of ~ 3.3 Å. This means that most of the similarity is due to the corresponding helices while the C or N termini in the Trp-cage and retro Trp-cage, respectively, differ significantly. More importantly the amino acid side chains forming the hydrophobic cores in both molecules are quite different. There is no proline-tryptophan motif in 2LUF that would interact with central tryptophan residue and be further stabilized by interaction with Y18 (Y3 in the Trp-cage). On the other hand, a completely novel stacking interaction between W15 and R5 emerges (Table 3-5). Such stabilization could be assigned to the charged guanidinium group of arginine and the aromatic heterocycle of the tryptophan. Such interactions are known to contribute significantly to the overall stability of a protein.
It is interesting to observe how the tryptophan indole ring interacts with the rest of the amino acid residues. The alignment of the tryptophan (W) and tyrosine (Y) side chains is not only essential for helix formation but also crucial for Trp-cage formation. Here, I also wanted to investigate the arrangement of those amino acids that forms strong interactions with the W and Y aromatic rings. The W and Y aromatic ring arrangement is also affected by proline residues present towards C-terminal end of Trp-cage. In Trp-cage, addition of extra alanines to the N-terminus stabilized the helices and the whole miniprotein [10]. The additional alanines also stabilise the helices leading to increased rigidity of the W and Y aromatic rings. It proved that W and Y can effect the helical content. In the retro Trp-cage the tryptophan indole ring moves further away from the poly-proline stretch and close to the arginine and aspartate residues. In the Trp-cage the N-terminal asparagines (N1) and C-terminal serine (S20) amino acids makes close interaction thus providing additional stability to the cage-like structures.11
Apart from the mutations, reversion of the Trp-cage forming (TC5b) peptide gave clear information regarding changes in inter-residual interactions.12 Temperature and solvent effects were studied to describe how they effected secondary structure forming motifs and eventually tertiary structure formation. The change in the direction of TC5b peptide increased electrostatic interactions and decreased hydrophobic interactions. The hydrophobic collapse brings PPII-helices part closer to the tryptophan indole ring. In this process, alpha helices and PPII-helices play the major role. This can lead to 310-helices and salt bridge formation in later steps that stabilize the Trp-cage.
The role of proline in 310-helices / beta-turn
In the retro peptide, the PPII-helix is at the N-terminus. The alpha helix is initiated immediately following P9 (Proline 9) leading to a longer alpha helix thus inhibiting the formation of a 310-helix . Formation of the extended helix can be explained by the P9 initiation and the presence of TFE. Normally if a proline is present it will make a bend and terminates helices. The Trp-cage peptide folding is an exception as the proline present in the middle forms a 310-helix / beta-turn like structure. In retro Trp-cage peptide, proline initiates formation of helix and is exposed to solvent. In mutant peptides, it forms a bend in this region.
We used 2D 1H-13C HSQC spectra chemical shifts of the prolines to reveal conformation of the prolines. In 2D 1H-13C HSQC spectra chemical shifts of the prolines show Trans conformation (Figure 4-11). This is similar to Trp-cage prolines. In the retro peptide, prolines are directed towards the outside, and are exposed to solvent while in the Trp-cage these are oriented towards the tryptophan side chain indole ring and form a hydrophobic core. The exposure of the PPII-helix proline to solvent suggests that the retro Trp-cage behaves like an unfolded protein.13 The 310-helix / beta-turn region differs in normal and retro Trp-cage peptides. The proline present in this 310-helix / beta-turn region is crucial for Trp-cage. Normally the conformation of proline is restricted due to the absence of free rotation around the nitrogen - α-carbon bond. Since the peptide bond nitrogen of proline lacks a hydrogen atom to contribute to a hydrogen bond, proline can only be stably accommodated with in the first turn of a α-helix. When present elsewhere, proline disrupts the conformation of the alpha-helix or beta-turn, producing a bend.
PPII-helices interaction with the α-helix
The sequence adopting the PPII conformation makes strong interactions with the helix part by trapping the tryptophan indole ring in the Trp-cage. In retro sequences, the proline region is poorly defined. There are only weak or moderate interactions observed between alpha and PPII-helix of retro and mutant Trp-cage peptides. Side chain residues around the tryptophan can affect the Indole ring which then can adopt a confirmation that avoids contact with PPII-helix prolines. The PPII-helix is quite flexible in retro Trp-cage peptide and its mutants compare to trp-cage. In fact, this part can be considered as an unfolded region of the retro Trp-cage peptide. Minor changes in the alpha helix part can disrupt PPII-helix. Favourable orientation of the amino acid residues in the PPII-helix and the α-helix region leads to successful cage formation.
The retro Trp-cage peptide showed a lower Tm than Trp-cage. The difference is approximately 10 °C (The retro Trp-cage peptide Tm measured in the presence of 30% TFE). The biphasic model of Trp-cage unfolding also supports opening of the PPII-helix and unwinding of the α-helix.16,17 The reason for Trp-cage unfolding at higher temperatures is the salvation of peptide side chains.18,19 Trp-cage, retro Trp-cage and its mutants show similarities in the α-helix and PPII-helix regions but differ in 310-helix formation. The relative orientation of tryptophan and proline side chains is completely reversed in the Trp-cage.
In the retro Trp-cage, 310-helix region is unstable and shows only turn formation. In the Trp-cage, this region adopts a stable conformation due to salt-bridge formation and proline (P12) ring pucker via stereo electronic effects.20 Salt-bridge formation may be a prerequisite for Trp-cage formation.21-23 In order for this to occur proper distance has to be maintained between arginine (R5) and aspartate (D12) residues of the retro peptides. Reversing the direction of the Trp-cage sequence revealed the importance of motifs in the Trp-cage fold. Structural analysis of the Trp-cage miniprotein showed how the altered amino-acid arrangements can lead to a different protein fold. peptide always folds so that it achieves the lowest possible energy. Heating the sample to 90 °C and cooling it down allows the peptide to get into a stable fold. No difference was found before and after the temperature refolding of retro peptide (Figure 12). This was in agreement with a description of retro peptide folding as obtained from computer models (in silico) or from experiments in the laboratory (in vitro) in which individual peptides were observed to be folding back to their original form following denaturation.
Assignment |
w1 |
w2 |
Data Height |
Note |
P3HBa-Y18QE |
1.610 |
6.416 |
117199.1 |
1 |
P4HA-Y18HBa |
4.056 |
2.725 |
943014.1 |
1 |
P4HA-Y18HBb |
4.057 |
2.643 |
931254.9 |
1 |
P4QG-W15HD1 |
1.652 |
6.949 |
83483.25 |
1 |
R5HBa-K13HN |
1.321 |
7.980 |
204203.4 |
1 |
R5HBb-W15HD1 |
1.427 |
6.950 |
198946.6 |
1 |
R5QD-K13QD |
2.719 |
1.325 |
334086.5 |
1 |
G10QA-W15HD1 |
3.607 |
6.951 |
454190.0 |
1 |
G10QA-W15HE3 |
3.607 |
7.222 |
457595.4 |
1 |
K13HN-R5HBa |
7.978 |
1.320 |
193054.8 |
1 |
K13QD-R5QD |
1.325 |
2.718 |
321554.4 |
1 |
W15HA-N20HBb |
4.215 |
2.480 |
883172.5 |
1 |
W15HE3-N20HA |
7.221 |
4.201 |
172554.9 |
1 |
Y18HBa-P4HA |
2.725 |
4.058 |
391550.6 |
1 |
Y18HBb-P4HA |
2.642 |
4.058 |
416109.7 |
1 |
N20HA-W15HE3 |
4.202 |
7.220 |
1022043 |
1 |
N20HBb-W15HA |
2.480 |
4.215 |
168045.3 |
1 |
Table 4 Starting set of long-range NOEs of the retro Trp-cage P9A mutant
Assignment |
w1 |
w2 |
Data Height |
Note |
S01HBa-Y18HA |
3.749 |
4.148 |
27435.2 |
l |
P02HBa-Y18QB |
1.705 |
2.971 |
1743352 |
l |
P04HA-W15HZ2 |
4.335 |
7.374 |
681809.9 |
l |
R05HA-W15HD1 |
4.153 |
7.225 |
672936.4 |
l |
R05HA-N20H |
4.153 |
7.686 |
504786.8 |
l |
R05HBa-Y18QB |
1.572 |
2.975 |
2237256 |
l |
R05HBb-W15HD1 |
1.698 |
7.223 |
618486.8 |
l |
R05QD-W15HE3 |
2.978 |
7.463 |
1142102 |
l |
R05QD-L19H |
2.978 |
8.205 |
2548468 |
l |
R05QG-Y18QB |
1.480 |
2.972 |
4222668 |
l |
S07HA-W15HD1 |
4.390 |
7.223 |
2311255 |
l |
S07HBa-W15HD1 |
3.851 |
7.226 |
710528.2 |
l |
S07HBb-W15HD1 |
3.784 |
7.226 |
723452.1 |
l |
S07HBb-W15HE3 |
3.784 |
7.465 |
859412.4 |
l |
S09HA-W15H |
4.388 |
7.872 |
3765239 |
l |
S09HA-Y18H |
4.387 |
8.102 |
1422784 |
l |
S09HA-L19MDa |
4.388 |
0.744 |
1117665 |
l |
L14HA-P03HDb |
4.044 |
3.649 |
759466.2 |
l |
L14HG-L19H |
1.582 |
8.206 |
3753458 |
l |
W15HE3-R05QD |
7.463 |
2.978 |
713955.7 |
l |
W15HE3-S07HBb |
7.462 |
3.784 |
923664.3 |
l |
W15HH2-N20H |
7.100 |
7.687 |
4263977 |
l |
W15HH2-N20HBa |
7.100 |
2.663 |
543359.8 |
l |
W15HH2-N20HBb |
7.101 |
2.721 |
682385.9 |
l |
Y18HA-S01HBa |
4.146 |
3.748 |
1174127 |
l |
Y18HA-P04HDa |
4.146 |
3.474 |
660192.8 |
l |
Y18QB-P02HBa |
2.974 |
1.707 |
1364514 |
l |
Y18QB-R05HBa |
2.974 |
1.574 |
2251971 |
l |
Y18QB-R05QG |
2.974 |
1.481 |
3485046 |
l |
L19H-R05QD |
8.201 |
2.978 |
1941777 |
l |
L19H-L14HG |
8.201 |
1.579 |
2864692 |
l |
L19HA-L14HG |
3.855 |
1.582 |
2778789 |
l |
N20H-W15HH2 |
7.686 |
7.099 |
3905116 |
l |
N20HA-W15HH2 |
4.469 |
7.099 |
1838242 |
l |
N20HBb-W15HH2 |
2.723 |
7.098 |
742635.6 |
l |
Table 5 Starting set of long-range NOEs of the retro Trp-cage P9S mutant

Figure 9 NMR structures of the Trp-cage (1L2Y (38 structures, RMSD 0.5 Ǻ), left and the retro peptide 2LUF (10 structures, RMSD 1.2 Ǻ), right. Prolines are shown in green, arginine in black, tryptophan in red and tyrosine in blue.

Figure 10 NMR structures of the retro Trp-cage mutants P9A (10 structures, RMSD 1.0 Ǻ), left, and the P9S (10 structures, RMSD 1.8 Ǻ), right. Alanine is shown in cyan, serine is shown in magenta, prolines are shown in green, arginine in black, tryptophan in red and tyrosine in blue.

Figure 11 The chemical shifts of the proline (showing trans configuration Cβ ~32 and Cγ ~27) in retro Trp-cage peptide.
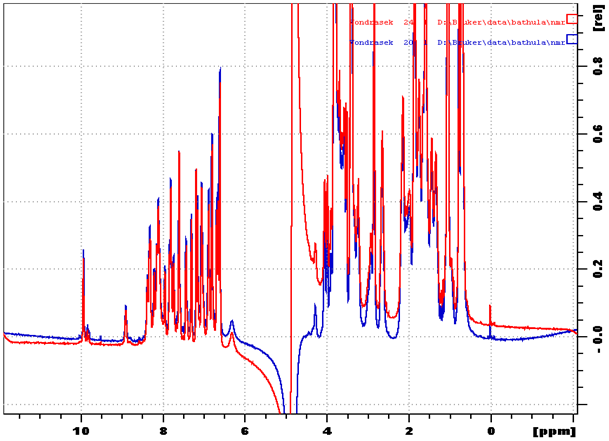
Figure 12 Figure shows proton chemical shift positions of retro peptide before temperature refolding (shown in red) and after temperature refolding (shown in blue) measured at 2 °C.
Temperature refolding of retro Trp-cage peptide.
MD simulations and fold prediction
MD simulations, Robetta and Pep Fold structure predictions were performed in collaboration with the group of Jiří Vondrášek from the Academy of Sciences of the Czech Republic. We determined experimental 3D structures. Analysis of similarities and differences between experimental structures and computer models and discussion of solvent effects are presented.24 The obtained NMR structure was also used to test the ability of computational tools to predict the correct fold of the retro Trp-cage. MD simulations, Robetta and Pep Fold modeling servers for automated structure prediction were unable to derive a structure similar to the NMR experimental structure. Addition of a mixed solvent system also did not help us in structure calculations.
This work discusses the more basic principles of Trp-cage miniprotein folding. Trp-cage structural study got attention due to its size and unique folding. In the cage formation, inter residual interactions play a crucial role. Using retro Trp-cage peptide we found the role of proline present on 310-helix is crucial for Trp-cage formation. Solvent effects were observed but they were secondary in Trp-cage formation. Structural analysis of small Trp-cage protein showed how the motif arrangement can lead to unique protein folding. Reversing peptide sequence from N-terminus to C-terminus can give us an idea about the flexibility and importance of independent motifs and their role in Trp-cage formation. A retro Trp-cage peptide and its mutants were studied to explain how rearrangement of motifs can lead to changes in secondary and quaternary structures. Results showed that, the direction of peptide had critical effect on amino acid like proline present in 310-helix /beta turn. This single proline can reform inter- and intra-residual interactions of whole peptide. The computational tools were unable to predict the correct fold of the retro Trp-cage. MD simulations, Robetta and Pep Fold modeling servers for automated structure prediction were failed to generate a structure similar to the NMR experimental structure. Addition of a mixed solvent system also did not help us in structure calculations. Our understanding related to protein folding is limited. We made an effort to prove that Trp-cage is vulnerable to retro operation. During our structural study of retro Trp-cage peptide and its mutants, the possibilities of cage formation were analysed.
Trp-cage can be easily affected by solvent and temperature. The protein secondary structure formation depends on its sequence as well as environment. The computational tools were unable to predict the correct fold of the retro Trp-cage. Although computing power continues to increase, our understanding related to protein folding is limited. Using retro Trp-cage peptide and its mutants we found that the role of the proline present on 310-helix is crucial for Trp-cage formation. Solvent effects were observed but they were secondary in Trp-cage formation. On the other hand tryptophan indole ring stacking interaction with the arginine guanidinium group contributes to the overall stability of retro Trp-cage peptides.
None.
The author declares no conflict of interest.

©2016 Bathula. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.