MOJ
eISSN: 2374-6920


Review Article Volume 6 Issue 2
1Rutgers Center for Integrative Proteomics, USA
2Biotech Support Group LLC, USA
Correspondence: Matthew Kuruc, Biotech Support Group LLC, USA, Tel 7322742866
Received: August 10, 2017 | Published: September 12, 2017
Citation: Zheng H, Roy S, Soherwardy A, et al. Stroma liquid biopsy™-a proteomic model of the systemic response to cancer. MOJ Proteomics Bioinform. 2017;6(1):236-241. DOI: 10.15406/mojpb.2017.06.00188
The concept of liquid biopsy has generated much scientific and commercial enthusiasm as it starts with an accessible body fluid, typically blood. These samples can then be monitored for example, to characterize the landscape of cancer-associated DNA mutations. These contributions notwithstanding, it is now overwhelmingly apparent that throughout cancer progression, there are adaptive microenvironments to support metastatic disease that are derived from the host systemic response. We now present evidence that host mediators between stroma and proliferating cells can in part, be monitored through the protein response that tracks into the vascularized tumor and re-proportions the extracellular proteins (serum) found in the general blood circulation. This rewiring pattern is especially significant, measurable even at very early stages of cancer, for many if not most primary tumors. This new multi-parameter cancer profile of progressive disease provides opportunities to monitor risk factors, for early detection, prognosis, recurrence, and guidance for therapeutic decisions.
Keywords: liquid biopsy, cancer biomarkers, serum proteomics, stroma liquid biopsy, cancer proteomic profile
The advancements in “next gen” and rapid genome sequencing have spawned new companies for liquid biopsy products and services that are quick, minimally invasive and that allow clinicians to monitor the course of therapy and to forecast recurrent disease. While these are certainly exciting prospects, challenges remain as genomic instability is a fundamental hallmark of cancer cells.1 So in many respects, genomic driven approaches will always suffer the problem of shooting after a moving target.
Likewise, discovery of protein biomarkers that can detect cancer early and personalize a treatment process has become an important research area in the proteomics field. For this, many proteomics approaches are being implemented in cancer research. These markers, foremost discovered through differential gene expression and shed from cancer cells, if at all measurable are of very low abundance (pg/ml range) in blood. To discover, characterize and monitor these ‘needle in the haystack’ biomarkers remains an industry wide challenge.
Beyond these inherent assay challenges, a tumor is more than simply a collection of immortalized cells. Cancer does not arise in a strictly mutational manner, but rather must be viewed as an entire ecosystem that also involves the individualized host response. As such, cancer cannot be fully characterized through the autonomous properties of only proliferating cells, without consideration for the supporting microenvironments.
The stroma1,2 Because of this, the current view of liquid biopsy is insufficient, as it relies solely through the analyses of the tumor cell genome and remnants from the cancer cells. It would therefore be highly desirable to have some additional window of observation that can monitor the supporting microenvironments – the stroma, yet collected and measurable from non-invasive sources, blood for example, to characterize progressive disease. We report here one such characteristic model for this purpose.
The stroma is a diverse milieu of soluble and membrane-bound proteins mediating multiple cellular components and biological processes. Reciprocal communication between tumor and stromal cells regulates the diverse components of the extracellular matrix, ultimately promoting tumor growth, survival and eventual colonization to metastatic sites3. So certain tissue microenvironments (the soil) may be especially hospitable to early disease or to new metastatic lesions. Consequently, metastatic potential is likely to depend upon some of the same supportive mechanisms in the microenvironments of the stroma (the soil) for the hospitality needed for reseeding and colonization by circulating cancer cells from the primary tumor1. The metastatic potential thus is forever dependent on the constitutional complexity of all the cellular and signaling interactions dynamically modified throughout tumor growth and metastasis. This concept is therapeutically important. As stromal cells within the microenvironments of tumors are genetically stable compared to tumor cells, their derivative proteomes may offer attractive therapeutic targets to help manage an often incurable disease and prolong survival.1,2 As such, any and all multi-parametric profiles that can help to monitor and stratify cancer patients for individual clinical situations will become highly desirable.3,4
We hypothesized that because of tumor vasculature, changes in the serum proteome might associate to the dynamic composition of the tumor-associated stromal microenvironments, and can thus be monitored by blood tests. We can now envision such measurements contributing to a Stroma Liquid Biopsy™ and we adopt this new term here to differentiate our model from previous liquid biopsies.
Liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS) provides and alternative assay approach to identify and measure proteins in an unbiased comparison ‘omic’ context. LC-MS/MS relies on the discriminating power of mass analyzers to observe specific trypsin-derived peptides as surrogates for the full gene product protein(s), and ion current intensities for quantitation. From discovery studies reported previously, we set out first to reduce the influence of albumin by incorporating AlbuVoid™ according to manufacturer’s instructions (Biotech Support Group LLC, Monmouth Junction, NJ USA), followed by LC-MS/MS analyses.5,6 These initial results were pared down to a target panel of serum protein markers, which when measured label-free by multiple reaction monitoring (MRM), provided evidence for a pattern or signature significantly different in the cancer patient population compared to an approximate sex/age matched normal and healthy control population. The special significance of this pattern is that these changes were categorical and primarily contained within these three host systemic response pathways: acute-phase inflammation, coagulation, and the complement cascade, illustrated on Figure 1.
Although some of these biomarker proteins have been previously described in the technical literature as potential biomarkers for cancer, the biomarkers within a pattern profile is not in the public domain and are patent pending.7–10 Even so, some of the biomarkers in the pattern are not based on the fully intact gene product, but rather truncated proteoforms resulting from proteolysis that can now be monitored by our methods. An important advantage of the use of biomarkers in our panel is that they are all highly observable with serum concentrations in the [µg-mg]/ml range, a range previously determined to be quantitative by LC-MS/MS with precision comparable to current clinical immunoassays.11 Most importantly, quantitative LC-MS/MS provides a singular platform for profiling multiple biomarkers in one analysis, using µl scale sample volumes.
Four normal female human samples were provided between the ages of 40 and 60, along with five cancerous serum samples from females within similar age range (Discovery Life Sciences, Los Osos, CA). The diseased samples were as follows: 1) stage 1 breast cancer, 2) stage 2 lung cancer, 3) stage 2b pancreatic cancer, 4) ovarian cancer, and 5) non-Hodgkin’s lymphoma cancer. The AlbuVoid™ Kit (Biotech Support Group LLC, Monmouth Junction NJ) was used to pre-fraction the samples to deplete albumin and simultaneously enrich the remaining sub-proteome on the bead. 50µl of each sample was individually processed with 25mg of AlbuVoid™ beads. The protocol provided in the kit was followed and samples were eluted with 200µl of the AlbuVoid™ Elution Buffer. The FASP method was used for reduction and digestion with modifications12. 20µl of eluate was added to the loading buffer (100mM Tris pH 7.6, 1% LDS (lithium dodecyl sulfate)) and 100mM DTT for reduction. Sample was incubated at 60C for 5min and centrifuged. Sample was added with 200µl of Urea buffer (8M deionized urea, 100mM Tris-HCL, pH 8.5) to a 30kDa Millipore filter (Amicon Ultra Cellulose) that was previously rinsed with urea buffer. Samples were spun at 10,000g for 15-20minutes, and filtrate discarded. Tube was washed with urea buffer and then alkylated with 22.5mM iodoacetamide, in urea buffer, and incubated for 40min in the dark at R.T. Tube was spun and washed with urea buffer 3 times, followed by with 50mM ammonium bicarbonate 3 times. Filter was transferred to new tube and 2ug of Pierce™ Trypsin Protease was added to digest the proteins. Tube was put on shaker for 10mins and left overnight in 37°C. Tube was spun to extract proteins and further rinsed with 75ul of 50mM ammonium bicarbonate. Filtrate was dried and re-solubilized to 40µl with 0.1%TFA.
Sample was analyzed by LC-MS using Nano LC-MS/MS (Dionex Ultimate 3000 RLSCnano System) interfaced with QExactive HF (Thermo Fisher, Waltham MA). Samples were loaded onto a self-packed 100µmx2cm trap (Magic C18AQ, 5µm 200 A, Michrom Bioresources, Inc.) and washed with loading Buffer A (0.1% trifluoroacetic acid) for 5min with a flow rate of 10µl/min. The trap was brought in-line with the analytical column (Magic C18AQ, 3µm 200 A , 75µmx50cm) and peptides fractionated at 300nL/min using a segmented linear gradient 4-15% B in 15min ( where A: 0.2% formic acid, and B: 0.16% formic acid, 80% acetonitrile), 15-25%B in 40min, and 25-50%B in 32min. Mass spectrometry data was acquired using a data-dependent acquisition procedure with a cyclic series of a full scan acquired in Orbitrap with resolution of 120,000 followed by MS/MS (HCD relative collision energy 27%) of the 20 most intense ions and a dynamic exclusion duration of 20 sec.
The raw data was converted into MASCOT Generic Format (MGF) using Proteome Discover 2.1 (Thermo Fisher, Waltham MA) and searched against Uniprot human database using an in-house version of X!tandem (Global Proteome Machine (GPM) software). For MS based quantitation, the raw data was analyzed using Skyline (Skyline-daily). The Skyline results were accepted if average mass error was below 3 ppm and the isotope dot product was greater than 0.9. For targeted MSMS, the target peptides were chosen based on search results and MS quantitation results. Retention time and M/Z of selected peptides were outputted from Skyline analysis and inputted as inclusion list into Xcalibur instrument method file for target MSMS. Samples were then analyzed again with the same LC-conditions using scheduled targeted MSMS with each target acquired for +/-2.5min of the peak retention time during the data-dependent run. AGC target was set at 5E5, resolution 30,000, maximum ion time 200ms, isolation window 1.4m/z, relative collision energy 27%. The LC-MRM-MS data were analyzed with skyline using the data-dependent runs as spectra library. Area of 6 best b or y ions was used for quantitation
The systemic response to cancer
On Table 1, we list a panel of proteins that we can monitor for three essential systemic responses to the presence of cancerous tissue. Because many of these have not been previously reported in multiple cancers, proteins reported as “undisclosed” remain proprietary. These protein markers are associated with 1) acute-phase inflammation, 2) coagulation, and 3) complement. The dysregulation reflects threshold differences between the normal/healthy population and the cancer population. These quantitative thresholds need to withstand the normal/healthy variance contributed from the combined biological and analytical variance.
From our results, though preliminary, the biomarkers chosen are exceedingly stable in the normal/healthy population while most are beyond the 2x arbitrary thresholds in the cancer population. Note that many of the biomarkers are severely re-proportioned in the cancer pattern. As the thresholds are preliminary, this report serves only to establish that three systemic response pathways are being dysregulated through progressive disease. The current thresholds are used foremost to illustrate this point. With more extensive sample types and data aggregation, we expect to optimize the biomarker methods and thresholds, to generate a much more defined pattern of cancer dysregulation for the variety of purposes proposed here.12
Furthermore, this study signifies the importance of these pathways intercommunicating in the vast circuitry of cascading proteolytic events, the predominant mechanism for controlling acute insults in the bloodstream. Because proteolysis is irreversible and therefore highly regulated, the pathways in our model cannot be viewed as separate independent cascades, but rather as one interdependent system with extensive cross-talk, mutually fine-tuning their functional status.13–15 Therefore new strategies to modulate proteolysis in the bloodstream may help unwind the triangulation of coordinated mechanisms in order to control disease progression and metastasis. In the sections which follow, we briefly highlight how some of these essential pathway interactions might play a role throughout cancer progression.
Acute-phase inflammation
The functional relationship between inflammation and cancer is not new and is now generally accepted that many cancers arise from sites of infection and chronic inflammation. Acute inflammatory diseases are usually self-regulating but persistent inflammation comes in play as a failure of mechanisms to resolve acute inflammatory response. It may be this inability to switch off the acute response that makes chronic inflammation a contributing factor in a variety of cancers despite it having many of the same mediators (e.g. cytokines and free radicals) as those generated during acute inflammation.16
One such special inflammatory case in our proteomic biomarker pattern is Alpha-1-Antitrypsin (AAT). Because of its unique characteristic as suicidal protease inhibitor belonging to the SERPIN gene super family, exhaustion of its inhibitory capacity may in part be one of the mechanistic failures to resolve the acute response and contribute to chronic inflammation. In a Review article in 2004, clinical oncologists, Drs. Sun & Yang described AAT as an alarming factor in malignancy and suggested there may be an imbalance between Alpha-1-Antitrypsin (AAT) & Neutrophil Elastase (NE) activities that could play a role in the progression of cancer.17
While beyond the scope of this report, using LC-MS reporting metrics, we now can observe peptide features of 2 variant sub-populations of AAT of opposite function; reporting either as an [inhibitory active proteoform] or an [inactive proteoform] (Table 2).
We find that the cancer sera signature generally follows a decline in the abundancy of [inhibitory active] AAT sub-populations relative to the total population. We suspect that in a normal and healthy population, there would be a sufficient blood reservoir of [inhibitory active] AAT. However, because of heredity, environmental risk factors, or progressive disease, in cancer populations, this reservoir becomes depleted, and the body can no longer replenish sufficient quantities of [inhibitory active] AAT to sustain inhibition of NE. Indeed if this model of dysregulation proves correct in vivo, it could contribute to a variety of therapeutic and detection strategies for the management and treatment of cancer.
Coagulation and complement cascade
While important, acute-phase inflammation however, is only part of the story. The coagulation and complement cascade are the other two legs of the Liquid Stroma Biopsy™ model. Patients with malignancy have a hyper-coagulable state due to the ability of almost all type of cancer cells to activate the coagulation system. Coagulation factors in cancer include the production of pro-coagulants directly from the tumor, as well as some general systemic responses of the host to the tumor, notably from inflammation and angiogenesis.18 That the coagulation system appears to be hijacked in support of cancer progression is further proof of a normal homeostatic function being dysregulated in cancer pathogenesis.
The third pathway in our model is the complement system, part of the innate immune system which in contrast to the adaptive immune system, does not change over the course of an individual's lifetime. It provides a powerful defense mechanism against pathogens as well as an endogenous danger sensor. However, dysregulated complement activation can have a significant role in both acute and chronic inflammatory conditions. Out of the three initiator pathways that activate the complement system, our cancer profile suggests that the initiators involved with the alternate complement pathway are diminished, while the terminal C5 remains unaltered. Though its role in cancer is complex and remains inconclusive, activated complement component proteins are abundantly dispersed throughout the inflammatory tumor microenvironment.19 Finally, there is overwhelming suggestion in the literature that induction of complement is beneficial for targeted monoclonal antibody cancer therapy.20
Protein conc. range > 5 log measurable in 1 LC-MS analysis |
Normal/Healthy females, Age 40-60 |
Cancer females, Age 40-60 |
|||||||||
Systemic pathway |
Appr. |
Uniprot gene identifier or |
N1 |
N2 |
N3 |
N4 |
Breast |
Lung |
N-hod |
Panc |
Ovary |
Coagulation |
100ng/ml |
CA (Undisclosed) |
Nd |
0.3 |
0.4 |
Nd |
0.8↑ |
3.2↑ |
4.0↑ |
1.0↑ |
2.5↑ |
Coagulation |
10µg/ml |
PPBP |
5 |
3 |
2 |
3 |
107↑ |
201↑ |
26↑ |
80↑ |
15↑ |
Coagulation |
10ng/ml |
CC (Undisclosed) |
Nd |
Nd |
Nd |
Nd |
39↑ |
87↑ |
11↑ |
22↑ |
11↑ |
Coagulation |
200ng/ml |
THBS1 |
0.1 |
Nd |
Nd |
Nd |
7↑ |
11↑ |
2↑ |
4↑ |
1↑ |
Coagulation |
80µg/ml |
CE (Undisclosed) |
1 |
1.4 |
0.9 |
1.5 |
0.6↓ |
0.5↓ |
0.3↓ |
0.2↓ |
0.2↓ |
Complement |
1,500 µg/ml |
TA (Undisclosed) |
1.8 |
1.3 |
1.6 |
1.3 |
1.2 |
0.5↓ |
1.1 |
0.5↓ |
0.5↓ |
Complement |
300µg/ml |
TB (Undisclosed) |
3.1 |
1.2 |
1.5 |
2.6 |
0.8↓ |
3.2 |
0.6↓ |
0.8↓ |
0.2↓ |
Complement |
25µg/ml |
TC (Undisclosed) |
2.5 |
1.6 |
1.3 |
2.3 |
1.2 |
1.5 |
0.6↓ |
0.8↓ |
0.4↓ |
Acute-phase Inflammation |
250ng/ml |
AB (Undisclosed) |
0.6 |
1.4 |
1.7 |
2.3 |
1.8 |
11.3↑ |
1.7 |
8.7↑ |
10.1↑ |
Acute-phase Inflammation |
5µg/ml |
SAA2 |
0.5 |
0.5 |
Nd |
0.4 |
0.8↑ |
15.2↑ |
4.8↑ |
4.9↑ |
1.0↑ |
Acute-phase Inflammation |
800ng/ml |
AD (Undisclosed) |
3.3 |
3.3 |
4.2 |
0.9 |
5.4 |
7.8↑ |
41.1↑ |
6.0↑ |
6.2↑ |
Table 1 LC-MS Quantification of Stroma Liquid Biopsy™ Protein Panel
The cancer sera samples were from patients with: stage 1 breast cancer, stage 2 lung cancer, stage 2b pancreatic cancer, ovarian cancer, and non-hodgkin’s lymphoma. Normal and cancer samples were acquired from a commercial source (Discovery Life Sciences, Los Osos, CA). Briefly, Mass spectrometry data was acquired at the Rutgers University Proteomics Center, using a data-dependent acquisition procedure with a cyclic series of a full scan acquired in Orbitrap with resolution of 120,000 followed by MS/MS (HCD relative collision energy 27%) of the 20 most intense ions and a dynamic exclusion duration of 20 sec. The raw data was converted into MASCOT Generic Format (MGF) using Proteome Discover 2.1 (ThermoFisher) and searched against Uniprot human database using an in-house version of X!tandem (Global Proteome Machine (GPM) software). For MS based quantitation, the raw data was analyzed using Skyline (Skyline-daily). For each protein, with the exception of CE, all spectral intensities were added together and reported as one number. The quantitation was normalized to a spiked reference standard. For CE, only one peptide feature was used as it held special significance as a regulating proteoform. To improve readability, all intensities were divided by 10, 100, 1000, or 10,000 as necessary to scale each row suitably. Thresholds were arbitrarily chosen as 2-fold increase or decrease from the average of the normals.
Nd Not detected ↑ Greater Than Threshold ↓ Less Than Threshold Bold numbers indicate severe dysregulation
Normal/healthy females, Age 40-60 |
Cancer females, Age 40-60 |
|||||||||
Column A |
Description |
N1 |
N2 |
N3 |
N4 |
Breast Stg 1 |
Lung Stg 2 |
N-Hod Lymph |
Panc |
Ovary |
sp|P01009|A1AT_HUMAN Total x100 |
SERPINA1 |
12.2 |
9.9 |
9.8 |
4.7 |
4.7 |
11 |
28.4 |
8.5 |
2.6 |
AAT RCL peptide x10000 |
GTEAAGAMFLEAIPMSIPPEVK |
6.8 |
10.8 |
7.5 |
1.5 |
0.1 |
0.5 |
4 |
0.4 |
0.1 |
Ratio: AAT/[AAT RCL] |
1.8 |
0.9 |
1.3 |
45.7 |
21.5 |
7.1 |
20.7 |
30.6 |
||
Table 2 LC-MS Quantification of AAT proteoforms
There are many different families of protease inhibitors, one exceptional family being the serine protease inhibitors, a collection of a super-family of proteins annotated within the SERPIN gene nomenclature. These proteinase inhibitors regulate key intracellular and extracellular pathways. Unlike more simple binary binding mechanisms, upon initial interaction with its target protease, SERPINs create a multitude of different and seemingly counter-intuitive opposing outcomes,. This is because SERPINs have a complex mechanism of action that involves a drastic change in their shape, forming the basis of a suicidal substrate inhibition mechanism and consequential sub-populations. As a result, functional regulation cannot be defined simply as an aggregate of all sub-populations, to differentiate serum phenotypes. Whereas this might be the case for non-SERPIN type inhibitors, an aggregate of all sub-populations does not account for important differences amongst the many sub-populations generated by conformational variants within this superfamily of proteins.
Observing and reporting the reactive centre loop (RCL)
The reactive centre loop (RCL) is a relatively short amino acid sequence which extends out from the body of the protein and directs binding to the target protease. The protease cleaves the serpin at the reactive bond site within the RCL, establishing a covalent linkage between the carboxyl group of the SERPIN reactive site and the serine hydroxyl of the protease. The resulting inactive serpin-protease complex is highly stable, and the structural disorder induces its proteolytic inactivation. As a consequence, the protease is permanently inhibited and functionally inactivated. Nevertheless, the story does not end there for the inhibitor, as after the initial interaction with the substrate protease, one of two possible outcomes can occur. One possible outcome is driven by covalent modification permanently inactivating the inhibitory capacity as the SERPIN peptide reactive bond region is irreversibly bound to the protease, and thus cannot be reconstituted back to an active form. The second possible outcome is a permanently inactive or “killed” proteoform of the SERPIN as the peptide RCL region is cleaved and can no longer bind target substrates. Our methods (same as described in Table 1) help to unravel sub-populations distinguishing an intact Reactive Centre Loop (RCL) region sub-population from a cleaved RCL region sub-population, distinguishing [inhibitory active] proteoforms of AAT from the total population of AAT. When the ratio of the total spectral intensities of all AAT peptides is considered in Column A (the total AAT population), relative to only the RCL peptide region GTEAAGAMFLEAIPMSIPPEVK, a severe pattern of dysregulation is observable in the cancer sera. This dysregulation pattern suggests that the [inhibitory active] proteoform of AAT is down-regulated or exhausted with progressive disease, regardless of stage or primary tissue of origin.
Cancer cells modify the surrounding microenvironments to serve its needs for nutrients, immune evasion, and space. Our observations corroborate other reports in the field that categorical mechanisms of coagulation, complement, and acute-phase inflammation are associated with cancer. Consistent with our model, a persistent inflammatory response observed in or around developing neoplasms has been shown to regulate many aspects of tumor development, from initiation all the way to metastatic progression.21
Notwithstanding these features, the cancer phenotype ultimately displays an individualized interplay between the host systemic response and the tumor. This individualized response is derived from a multitude of factors from the patient host not predicated directly upon the proliferating cancer cells, Figure 2. One important systemic response-the immune response is especially important as prolonged survival has been demonstrated in the clinic for immune checkpoint modulators like PD1. With these advancements comes the recognition that manipulation of the tumor microenvironment to enhance T cell infiltration and access to the tumor will be necessary to affect better long-term immunotherapy outcomes. This will require a more thorough and reportable characterization of the host immune response; the individualized process involving interactions between the tumor and microenvironment including multiple cell subsets and soluble mediators functioning at different times and at different anatomical sites within the tumor stroma and vasculature.
Yet, despite the vast amount of research on genetic tumor profiling, to the best of our knowledge, somatic mutations that predict response to immunotherapy do not exist. Alternative biomarkers are necessary. We solicit that there is great advantage to the variety of highly observable and quantifiable proteins that form a serum pattern of normalcy in a healthy person without cancer. By contrast this same panel of proteins is dysregulated by the localized subversion of normal homeostatic mechanisms within the tumor microenvironments. Such dysregulation forms a cancer-specific serum pattern of great novelty; that of activation of acute-phase inflammation and coagulation, and de-activation of the early initiators of the complement cascade. Our data suggests that this pattern is measurable even at very early stages of cancer, for many if not most primary tumors. Therefore it reflects in part, the individualized density and soluble mediators of the cellular composition of the tumor stroma. Consequently, using proteomic data from the proposed panel here, we envision to match patients most likely to benefit and least likely to experience adverse events, and ideally to monitor the change in phenotypes upon treatment, to establish long-term responsiveness and survival benefits for immuno-oncology and related therapies, all with relatively easy, non-invasive measurements. In much the same manner, methods to monitor new therapeutic strategies to modulate interconnected proteolytic events and to manipulate the tumor microenvironment for therapeutic effect, may also be established.
This report serves only as an introduction to the prospects for compiling Stroma Liquid Biopsy™ proteomic data. In future investigations, we hope to refine these reporting metrics with large cohorts of samples, so we might find patterns for early cancer dysregulation and response to therapies, regardless of the primary tumor. Under the guidance of a physician, the concept for Stroma Liquid Biopsy™ might then be a realistic goal. It would generate objective measures whether or not to rule out cancer as a possible diagnosis when risk factors are accessed, or even screen for early detection. Tissue specific factors may add further discretion into this panel. Though not considered in this report, several biomarkers mostly derived from the same SERPIN gene super family as AAT, do seem to be more or less dysregulated within the 5 primary tumors tested to date, data not shown. Whether all these patterns correlate best with primary tissue of origin, or staging or some combination of tissue and staging, needs further investigation. Likewise co-pathologies and other chronic conditions which may impose changes to the proteins in this panel needs further investigation.
Therefore, it is clear that before having any clinical utility is established, a large aggregation of data and associated characterization of cancer is required. Yet such data will be necessary as complementary to data adopted from other Liquid Biopsy analyses. Data gathering platforms based on nextgen sequencing, circulating tumor cells, exosomes, or more conventional tumor burden biomarkers (i.e. CEA) are limited now, and may ultimately always be limited to monitor the host systemic response to the presence of cancerous tissue. Monitoring such host response is the opportunity presented by our Stroma Liquid Biopsy™ panel of biomarkers and characteristic pattern of disease. Potentially any oncology therapy that can unwind the interconnections between the three systemic response pathways reported here and promote a more
Normal/healthy characteristic pattern, can find purpose to adopt a companion Stroma Liquid Biopsy™ biomarker panel. Thus, Stroma Liquid Biopsy™ panels can serve many medical applications in the detection, management and treatment of cancer.
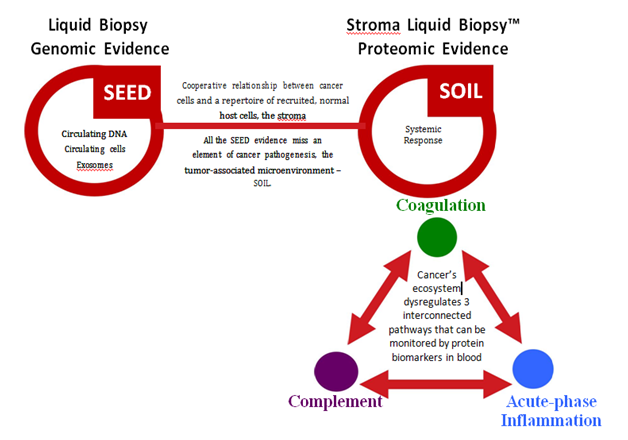
Figure 2 Stroma Liquid Biopsy™ Reports Dysregulation of 3 Interconnected Systemic Pathways in Cancer.
The localized microenvironments of tumors cooperate to subvert normal homeostatic mechanisms in blood, re-proportioning a panel of serum proteins, individualized as patterns, but consistent systemically with the presence of cancer.

©2017 Zheng, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.