MOJ
eISSN: 2374-6920


Research Article Volume 5 Issue 2
1Department of Food Science, Zhejiang Pharmaceutical College, China
2College of Food Science and Technology, Nanjing Agricultural University, China
Correspondence: Zhifang Yu, College of Food Science and Technology, Nanjing Agricultural University, Nanjing, 210095, P.R. China, Tel ,, Tel +86 25 84399098, Fax +86 25 84395618
Received: January 12, 2017 | Published: March 7, 2017
Citation: Luo H, Wang L, Jiang L, et al. Responses of dynamic proteome changes in Zizania latifolia to fresh-cut operation during room temperature (25°C) storage. MOJ Proteomics Bioinform. 2017;5(2):55-64. DOI: 10.15406/mojpb.2017.05.00155
In order to explore the molecular mechanism of fresh-cut operation accelerating senescence and quality deterioration of Z. latifolia, the response of dynamic proteome change in Z. latifolia to fresh-cut operation during room temperature (25°C) storage was studied. 35 protein spots were identified to change significantly (p<0.05) from the total 660 protein spots on the gels by MALDI-TOF/TOF. Compared with the control, 29 proteins showed similar expression pattern after fresh-cut operations in spite of the difference in abundance between two treatments. Meanwhile, six spots (plant basic secretory protein, adenosine kinase, C2 domain, class III peroxidases, LbH_gamma_CA_like, small Ras-related GTP-binding protein), which were down-regulated in whole Z. latifolia, increased after fresh-cut operation during storage. Fresh-cut operations that accelerate senescence and quality deterioration of Z. latifolia might be due to the production and transduction of wounding signals, acceleration of carbohydrate and nucleotide catabolism, disorder of energy homeostasis, enhancement of free radical damage, as well as degradation of cell structure.
Keywords: zizania latifolia, fresh-cut, senescence, quality deterioration, proteomics
Zizania latifolia (Griseb.) Turcz. ex Stapf, botanically different from the three species (Z. aquatica, Z. palustris and Z. texana) grown in North America, is a perennial aquatic grass belonging to tribe Oryzeae of the grass family (Poaceae) in China.1 Z. latifolia grows along the littorals of freshwater lakes, streams, marshes and pools.2 It is usually parasitized by the smut fungus (Ustilago esculenta P. henn.), which stimulates enlargement of the culms of the grass.3,4 The enlarged infected culms have been eaten as a vegetable in China since the l0th century, commonly called Jiaobai, Jiaosun, Gusun, Gushou or Gaosun.5 Because of its highly nutritional and economic value, the vegetable has been widely cultivated in rice fields of south China.
Z. latifolia is very suitable to be processed into fresh-cut product. It is usually cut into a cube, slice or shred before marketing.6 However, fresh-cut operations may accelerate the senescence and quality deterioration triggering cut surface browning, tissue lignification, shriveling, microbial contamination, off-flavor, respiratory rate, cellulose and lignin contents increase, which reduced the shelf life of fresh-cut Z. latifolia to 2-3days at room temperature.6 Therefore, clarification of the mechanisms involved in senescence and quality deterioration is of great importance to extend the shelf life of fresh-cut products. Previously, the physiological and biochemical responses in Z. latifolia after fresh-cut operation have been investigated.7,8 In general, fresh-cut processing operations destroys the integrity of Z. latifolia, which in turn promotes a faster physiological deterioration and biochemical changes such as respiratory activity, ethylene production, reactive oxygen species and malondialdehyde accumulation, lignification and antioxidant-related enzymes changes etc., thus accelerate the rate of senescence and quality deterioration.7 Nevertheless, these researches are mainly focused on wounding stress-induced changes at physiological and biochemical levels.9
Fresh-cut products, which are subjected to mechanical damage by cutting operations, are still living organisms. Proteins play crucial roles in the structure and function of all living cells and together with nucleic acids, carbohydrates and lipids, form the biochemical basis of life.10,11 Currently, proteomic approaches are being extensively used in fruits and vegetables to gain understanding on the mechanisms related to ripening and regulation,12 senescence,13 stress response14 and preservation.15,16 Luo et al.,13 analyzed the protein profile in Z. latifolia during storage at 1°C and found that the senescence of postharvest Z. latifolia may be a consequence of multiple reasons, comprising the regulation of material metabolism, change of energy metabolism pathway, decline of reactive oxygen scavenging capacity and degradation of cell structure. In addition, we analyzed the changes in the protein profile of Z. latifolia during refrigerated (1°C) storage. Results showed that 25 proteins differentially expressed and identified, with their functions mainly concentrated on cell structure, stress response and defense, and senescence.14 However, the molecular mechanism of senescence and quality deterioration in fresh-cut Z. latifolia is still not well understood. Also, there is limited information available on the wounding-responsive proteins occurring in the vegetable tissues after fresh-cut operation during storage at room temperature.
The objective of this study was to determine the response of dynamic proteome change in Z. latifolia to fresh-cut operation during room temperature (25°C) storage. We hope our result would explore the molecular mechanism of fresh-cut operation accelerating the senescence and quality deterioration of Z. latifolia.
Plant material
Fresh Z. latifolia at mature stage were hand-harvested from a commercial farmland in Yixing, Jiangsu, China. 18Kg Z. latifolia with uniform size and invisible defects, were selected, rinsed with tap water, and then randomly divided into two groups. The first group was manually peeled and cut into slices of 3~5mm thickness with a sharp stainless steel knife, and the second group (whole stem) acted as the control. The slices and the whole stem (control) were then packaged into polyethylene plastic bags (thickness: 15μm, size: 40cm×60cm) before storage at 25°C. Samples (3Kg) were taken after 0, 3 and 5days, respectively, frozen in liquid nitrogen, and then stored at -20°C until further analysis.
Protein extraction and two-dimensional electrophoresis
Protein extraction was carried out according to the method of Liu et al.,7 10g sample ground into powder in liquid nitrogen was homogenized with 15ml of ice-cold solubilisation buffer (pH 8.3) consisting of 1mmolL-1 PMSF, 60mmolL-1 Tris, 1.05molL-1 sucrose, 10mmolL-1 EGTA, 1% Triton X-100 and 1mmolL-1 DTT. After centrifugation at 16,000×g for 30min at 4°C, the supernatant was collected and immediately mixed with three volumes of ice-cold Tris-saturated phenol (pH 7.8) followed by incubation at 4°C for at least 2h and centrifugation at 16,000×g for 30min. The upper phenolic phase was collected and precipitated overnight with five volumes of chilled acetone at -20°C prior to centrifugation at 16000×g for 30min at 4°C and rinsed twice with ice-cold methanol and acetone. The pellets were air dried at room temperature and dissolved overnight at 4°C with lysis buffer (7molL-1 urea, 2molL-1 thiourea, 4% (w/v) CHAPS, 1% (w/v) DTT and 0.5% (v/v) pH 4-7 IPG buffer). Protein concentrations were determined using Coomassie brilliant blue (CBB) G250 staining method, and samples were stored at -20°C until two-dimensional electrophoresis (2-DE).14
2-DE
1mg protein samples were diluted in a final volume of 350μL and loaded onto 17cm pH 4-7 immobilized pH gradient (IPG) strips (Bio-Rad). The running conditions were as follows: 50V for 12h (rehydration), 100V in slow for 1h, 200V in slow for 1h, 500V in slow for 1h, 1000 V in slow for 1h, 4000V in linear for 2h, 8000V in linear for 2h and then run at 8000 V for 7.5h. The strips were equilibrated for 15min in 2% (w/v) DTT with equilibration buffer (50mmolL-1 Tris-HCl pH 8.8, 6 molL-1 urea, 20% (v/v) glycerol and 2% (w/v) SDS) followed by 15min in 2.5% (w/v) iodoacetamide in equilibration buffer. For the second dimension, the focused strips were loaded and run on SDS-PAGE 12% polyacrylamide gels using the Ettan Six vertical set (GE Healthcare) for 1.5h at 1W/gel, followed by 15W/gel during 4h. After SDS-PAGE, gels were stained with 0.12% CBB G250. Three biological replicates were performed for each storage time.
Image analysis
The gels were scanned using a Versdoc 3000 scanner (Bio-Rad) at 300 dpi and saved as the GSC image files. Image analysis of each gel was performed using the PDQuest 2-DE analysis software Version 8.0.1 (Bio-Rad).
Tryptic digestion of spots
Spots showing changes statistically significant (at p<0.05) and above a 2-fold threshold were carefully excised from the gels. The protein spots digestion with trypsin was performed according to the method described by Luo et al.14
Identification of proteins by MALDI-TOF/TOF and database search
Protein identification was performed by Shanghai Bo-Yuan Biological Technology Co., LTD, Shanghai, China. Database search were performed using GPS Explorert™ software v3.6 (Applied Biosystems, USA) over National Center for Biotechnology Information nonredundant database using the MASCOT 2.1 (Matrix Science Ltd., London). The database search was restricted to green plants. The functional classification of the identified proteins was annotated using the database at http://uniprot.org/uniprot. For protein accessions identified by MASCOT that had no functional annotation, a function was assigned via BLASTP searches of the corresponding protein accessions.
2-DE analysis of whole and fresh-cut latifolia during storage at 25°C
During storage, the symptoms of senescence and quality deterioration in both whole and fresh-cut Z. latifolia are gradually severe including shell etiolation, surface browning, and tissue hollowness in whole Z. latifolia as well as cutting surface browning, and tissue lignification in fresh-cut Z. latifolia (Figure 1). The proteins from whole and fresh-cut Z. latifolia during storage were extracted and separated by 2-DE. The 2-DE maps were obtained using isoelectric focusing (IEF) on 17cm pH 4-7 IPG gels (Bio-Rad) followed by SDS-PAGE on 12% polyacrylamide gels before staining with Coomassie brilliant blue G250 (Figure 2). The 2-DE gels were matched and analyzed by PDQuest 2-DE analysis software Version 8.0.1. Approximately 660 protein spots were detected on the gels. Quantitative image analysis of three replicates of each sample revealed that 39 protein spots showed a more than 2.0-fold increase/decrease in protein abundance with p<0.05 in at least one storage time point compared to the whole Z. latifolia of corresponding time point. The specific distribution in Figure 3 and the magnified views of some differentially expressed proteins were shows in Figure 4.
Identification by mass spectrometry and functional analysis of differentially expressed proteins
35 out of 39 spots which significantly changes (p<0.05), were successfully identified and could then be classified into seven functional categories as listed in Figure 5, including signal transduction (8.6%), metabolism (22.9%), cell structure (8.6%), stress response and defense (31.4%), protein synthesis (11.4%), senescence (5.7%), and unclear functional proteins (11.4%). Cluster analysis was carried out on differentially expressed proteins from day 0 to day 5 (Figure 6), revealing a dynamic proteome profile shift of Z. latifolia at different time points during storage at 25°C.
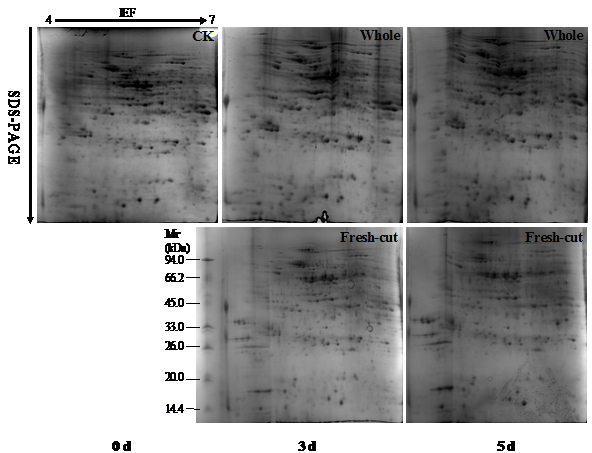
Figure 2 Representative 2-DE gels from whole and fresh-cut Z. latifolia during storage at 25 °C. 0 d was used to build the master gel by matching the 2-DE images of different storage time (3 and 5 d). Proteins (1mg) were separated on a 17 cm IPG strips (pH 4-7) and subsequently on a 12% SDS-PAGE gel. The spots were visualized by CCB G250 staining.
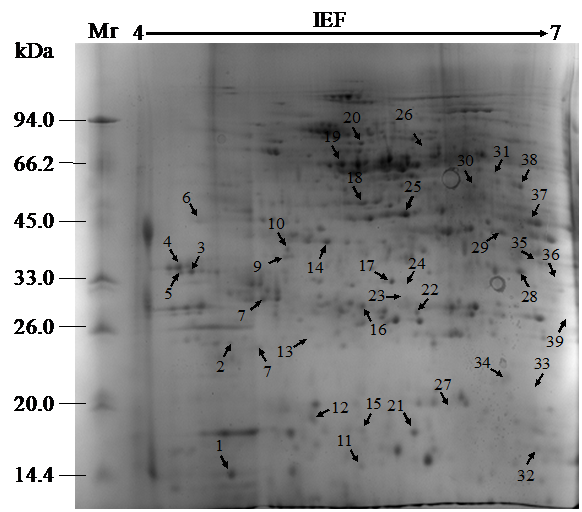
Figure 3 2-DE reference map of Z. latifolia. All differentially expressed proteins are numbered and indicated by arrows.
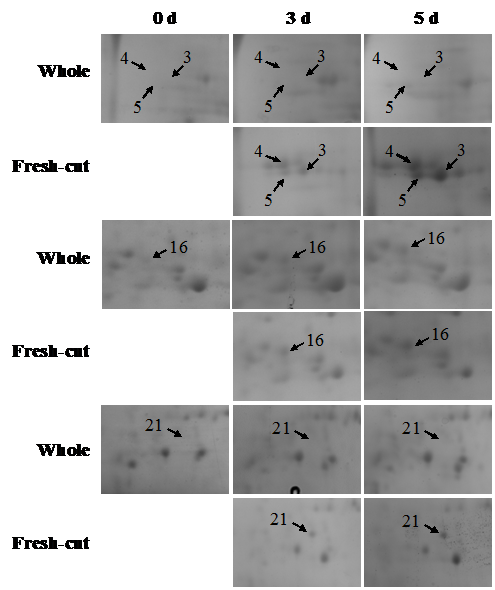
Figure 4 Magnified views of some differentially abundant proteins marked in Figure 2.
Different storage times were showed on the top. Arrows and numbers are indicating the differential spots among different storage time.
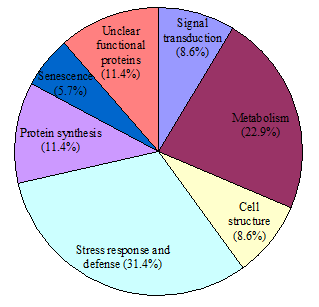
Figure 5 The functional distribution of identified proteins expressed differentially covering seven .
protein groups categorized according to their putative functions.
The numbers and percentages of each protein group are indicated.
As shown in Table 1, 14 out of the 35 differentially expressed and identified proteins were up-regulated while the other were down-regulated during the storage of whole Z. latifolia. For the up-regulated proteins, spot 14 (fructokinases), spot 17 (glycosyl hydrolases family 17), spot 19 (2,3-bisphosphoglycerate-independent phosphoglycerate mutase) and spot 26 (diphosphonucleotide phosphatase 1) were related to metabolism; spot 1 (profilin) to cell structure; spot 3 (beta-1,3-glucanase precursor), spot 4 (glucan endo-1,3-beta-glucosidase), spot 5 (beta-1,3-glucanase precursor), spot 12 (universal stress protein family), spot 21 (thaumatin-like protein isoform 2), spot 22 (glutathione S-transferase) and spot 28 (β-1,3-glucanase) to stress response and defense; spot 16 (cysteine protease) and spot 23 (papain-like cysteine proteinase) to senescence.
For the down-regulated ones, spot 8 (14-3-3 protein homologues), spot 10 (C2 domain) and spot 38 (small Ras-related GTP-binding protein) were related to signal transduction; spot 6 (adenosine kinase), spot 20 (transketolase), spot 30 (enolase) and spot 37 (isocitrate dehydrogenase (NADP+)) to metabolism; spot 18 (actin) and spot 27 (Actin depolymerisation factor/cofilin -like domains) to cell structure; spot 2 (plant basic secretory protein), spot 24 (class III peroxidases), spot 33 (dehydroascorbate reductase) and spot 34 (chitinase class I) to stress response and defense; spot 11 (glycine-rich RNA-binding protein), spot 13 (putative chaperonin 21 precursor), spot 15 (putative ribosomal protein S12) and spot 31 (RNA recognition motif) to protein synthesis; Furthermore, the functions of spot 9 (DREPP plasma membrane polypeptide), spot 29 (WD40 domain), spot 35 (LbH_gamma_CA_like) and spot 36 (ricinB_lectin_2) were still unclarified.
On the other hand, when compared to control tissue the responses of 29 proteins during storage were enhanced after fresh-cut operations. However, the expressions of spot 2, spot 6, spot 10, spot 24, spot 35, and spot 38, which decreased during storage of whole Z. latifolia, were up-regulated after fresh-cut operation, indicating that they might play vital roles in response to wounding stress.
During senescence and quality deterioration of Z. latifolia, a variety of physiological and biochemical responses occurred, in association with the changes in the protein profile, which could be influenced by a variety of internal or external factors could impact such process. In this study, changes in abundance of 35 proteins between whole and fresh-cut Z. latifolia during storage were observed based on 2-fold difference. They are suggested to play roles in signal transduction, carbohydrate and nucleotide metabolism, cell structure, stress response and defense, protein synthesis, as well as senescence.
Identified proteins related to signal transduction
Signal transduction is the process which begins with the binding of specific ligands to receptors located at the surface of the plasma membrane, thus causing an intracellular metabolic change.17 Three proteins involved in signal transduction were identified, including 14-3-3-like protein (Os08g0430500, spot 8), C2 domains (Os08g0562600, spot 10) and small ras-related GTP-binding protein (spot 38). 14-3-3-like proteins are important regulators of signal transduction pathways through binding to multiple functionally diverse signaling proteins such as kinases, phosphatases and transmembrane receptors.18,19 C2 domains are usually Ca2+-dependent membrane-targeting module found in many cellular proteins, functions in membrane trafficking or signal transduction.20 As a signaling element, small ras-related GTP-binding proteins meditate various external stimuli to intracellular signaling pathways.21,22 All of them decreased during the storage of whole Z. latifolia; however, spot 10 and spot 38 were up-regulated by fresh-cut operation, suggesting that signal transduction was enhanced.
Identified proteins related to metabolism
Regulation of physiological metabolism is an important strategy for plant to adapt to (a) biotic stresses.23 Two protein involved in nucleotide metabolism were identified: adenosine kinase (AK, spot 6) which is involved in the purine salvage pathway plays a key role in the prevention of adenosine accumulation and support the metabolic cycling of adenosine,24 while diphosphonucleotide phosphatase 1 (DPP, spot 26) catalyzes the hydrolysis of a wide range of activated phosphoric acid mono- and di-esters and anhydrides at pH of 5-6.25 In this study, the biosynthesis of AK and DPP increased after fresh-cut operation at 3rd and/or 5thdays of storage, indicating that nucleotide catabolism was enhanced.
Fructokinases (Os08g0113100) (FRKs, spot 14), glycosyl hydrolases family 17 SORBIDRAFT_09g018750) (GHF, spot 17), 2,3-bisphosphoglycerate-independent phosphoglycerate mutase (Os05g0482700) (BIPM, spot 19), transketolase (TK, spot 20), enolase (spot 30) and isocitrate dehydrogenase NADP+ (IDH, spot 37) play important roles in carbohydrate metabolism.26 Of these, GHF belong to the glycosyl hydrolase family and catalyzes the hydrolysis of polysaccharides to monosaccharides.27 Fresh-cut significantly increased the expression of GHF during storage, suggesting an enhanced carbohydrate catabolism. FRKs, BIPM and enolase are involved in Glycolysis,28 IDH is the key enzyme in the tricarboxylicacidcycle acid (TCA) cycle,29 while TK is the key enzyme of the pentose phosphate pathway.30 Glycolysis, TCA cycle and pentose phosphate pathways provide energy and inter-conversion building blocks for metabolites synthesis.11,15 The higher expressions of these proteins after fresh-cut operation implied that the reduction of energy supplement.
Identified proteins related to cell structure
The plant cell wall is of great importance to provide shape to cells, form the interface between adjacent cells as well as resisting the invasion of pathogens.31 Three proteins related to cell structure were identified, including profilin (Os06g0152100, spot 1), actin (spot 18) and actin depolymerisation factor/cofilin -like domains (Os03g0780400) (ADF, spot 27). Actin is a ubiquitous protein involved in the formation of filaments, a major component of the cytoskeleton.29 Profilin may link the cytoskeleton with major signaling pathways by interacting with components of the phosphatidylinositol cycle and Ras pathway, which plays a central role in the assembly of branched actin filament networks.32 On the other hand, ADF, a family member of essential eukaryotic actin regulatory proteins, could enhance the turnover rate of actin and interact with actin monomers and actin filaments.33 In comparison with those in control, the abundance of ADF was higher in fresh-cut Z. latifolia during storage while the biosynthesis of profilin and actin was down-regulated, suggesting that fresh-cut accelerated the cell structure disassembly, which was further validated by cell ultrastructural observation using transmission electron microscope (unpublished data).
Identified proteins related to stress response and defense
Stress response would cause alternation in composition of plant transcriptome, proteome and metabolome, which mediated via profound changes in gene expression.34 Of the eleven stress response and defense proteins up-regulated by fresh-cut operation, eight were related to disease resistance. plant basic secretory protein (BSP, spot 2) is believed to be part of the plants defense mechanism against pathogens,35 β-1,3-glucanase precursor (spot 3, 5), glucan endo-1,3-beta-glucosidase (spot 4) and β-1,3-glucanase (spot 28) could degrade fungal cell wall polysaccharides,36 universal stress protein family (USP, spot 12) is a small cytoplasmic bacterial protein whose expression is enhanced when the cell is exposed to stress agents,13 thaumatin-like protein isoform 2 (TLP, spot 21) plays a roles in host defense and developmental processes,37 while chitinase class I (spot 34) catalyzes the hydrolysis of the beta-1,4-N-acetyl-D-glucosamine linkages in chitin polymers of fungal cell walls.38 The higher abundances of these proteins (except chitinase class I) in fresh-cut Z. latifolia when compared to those in control provide a good indication that fresh-cut operation induced elevation in anti-pathogen capability.
Senescence is a vital aspect of vegetable life cycles, and directly affects vegetable quality and resistance to pathogens. Reactive oxygen species (ROS), as the primary mediators of oxidative damage in plants, are involved in senescence.39 Furthermore, superoxide dismutase (SOD), ascorbate peroxidase (APX), dehydroascorbate reductase (DHAR), glutathione S-transferase (GST) and peroxidases (POD) are antioxidant enzymes related to ROS metabolism.40 In this study, three ROS metabolism related proteins identified were GST (spot 22), class III peroxidases (PODIII, spot 24) and DHAR (spot 33). GST is cytosolic dimeric proteins involved in cellular detoxification by catalyzing the conjugation of glutathione (GSH) with a wide range of endogenous and xenobiotic alkylating agents,12 while DHAR catalyzes the reduction of dehydroascorbate into ascorbic acid (AsA) using GSH as the reductant.,40 Both GST and DHAR are involved in the ascorbate-glutathione cycle (Figure 7), which has been regarded as one of the most important antioxidant pathways for hydrogen peroxide (H2O2) detoxification. PODIII, located in the extracellular space or in the vacuole in plants, is involved in indole-3-acetic acid degradation, lignin biosynthesis, wound healing, general stress response and pathogen defense, and H2O2 detoxification.29 In previous studies, we found that fresh-cut significantly (p<0.05) promoted the increase of POD activity in Z. latifolia during 25°C storage (unpublished date), which was validated by POD expression pattern in the present study. Based on these results, we speculated that ROS damage could be involved in senescence and quality deterioration of fresh-cut Z. latifolia.
Identified proteins related to protein synthesis
The regulation of protein synthesis plays a key role in the control of cell growth, proliferation, and apoptosis.41 In this study, four proteins identified are responsible for protein synthesis, glycine-rich RNA-binding protein (GRP, spot 11), putative chaperonin 21 precursor (C21P, spot 13), putative ribosomal protein S12 (RPS12, spot 15) and RNA recognition motif (RRM, spot 31) were identified. GRP suggested to play a role in responses to environmental stresses,42 C21P help nascent peptide chain precursor reconciliation folded protein peptide chain folds into a protein possessing a biological function,43 RPS12, located at the subunit interface, supposedly play critical roles in interacting with the tRNA substrates and the large subunit,44 RRM, a highly abundant domain of the proteins in eukaryotes, participates in post-transcriptional gene expression processes, including mRNA and rRNA processing, RNA export, and RNA stability.45 The biosynthesis of these proteins was down-regulated after fresh-cut, suggesting that fresh-cut operation weaken the protein assimilation of Z. latifolia.
Identified proteins related to senescence
Senescence caused by pathogens, environmental stresses, or inherent physiological changes is a collective term for the deteriorative changes in living organisms, leading to death. Two proteins, cysteine proteases (CPs, spot 16) and papain-like CPs (spot 23), were supposed to be related with the senescence. CPs are enzymes that degrade proteins by cleave the peptide bond, and participated in a wide range of physiology and development processes.46 In this study, CPs (spot 16) and papain-like CPs (spot 23) significantly up-regulated after fresh-cut operation during storage. It is likely that the wounding stress caused by fresh-cut enhanced the protein catabolism, which may play a major role in senescence and quality deterioration of Z. latifolia.
The unclear functional proteins
Four spots identified as DREPP plasma membrane polypeptide (spot 9), WD40 domain (spot 29), LbH_gamma_CA_like (spot 35) and RicinB_lectin_2 (spot 36). Although they were searched by BLASTP matches of the corresponding protein accessions, their functions were still unclear. The expressions of spot 9, 29, and 36 showed similar expression pattern in both whole and fresh-cut Z. latifolia during storage; nevertheless, LbH_gamma_CA_like (spot 35), which decreased in whole stem, showed an opposite trend in fresh-cut tissue during storage. These results suggested that LbH_gamma_CA_like might play directly/indirectly role in response to wounding stress.
This study demonstrated that when compared to integral stem the dynamic proteome changes of 35 proteins identified were significantly altered after fresh-cut operation in association with the accelerated senescence and quality deterioration. Based on their functions, the possible mechanisms of fresh-cut operation accelerating senescence and quality deterioration of Z. latifolia might be due to: (1) enhancement of production and transduction of wounding signals; (2) enhancement of the carbohydrate, nucleotide and protein catabolism, but attenuation of the protein assimilation, glycolysis, TCA cycle and pentose phosphate pathways, thus reducing the energy supplement; (3) inducement of the expression of pathogenesis-related proteins and antioxidant enzymes; (4) acceleration of the cell structure disassembly. This study has put forward interpretable data and provided new perspectives on the mechanism of fresh-cut operations accelerating senescence and quality deterioration of Z. latifolia. However, further detailed investigation of these proteins’ specific roles and their functional correlation with wounding response should be performed to better understand the mechanism of fresh-cut operations accelerate senescence and quality deterioration.
This work was supported by the National Natural Science Foundation of China (31401612), the Natural Science Foundation of Zhejiang Province of China (LY14C200005), and the Zhejiang Open Foundation of the Most Important Subjects (KF2012006).
The author declares no conflict of interest.

©2017 Luo, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.